Hello,
I’m preprocessing multi-echo data in fMRIPrep with the goal of using tedana for denoising (on the individual echo images as provided by --me-output-echos). I am running both fMRIPrep 21.0.2 and Tedana 0.0.12 via Singularity on our HPC.
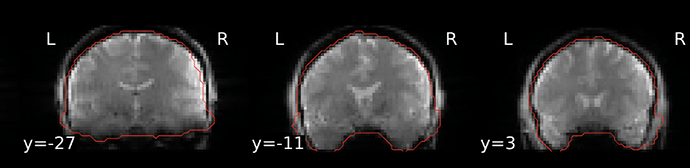
A number of the fMRIPrep functional reports show an overly-inclusive BOLD mask that incorporates areas of skull and dura (see example at bottom of post). Anatomical images and coregistration look good. The final fMRIPrep-derived optimally-combined image also looks good; skull-stripping appears accurate there.
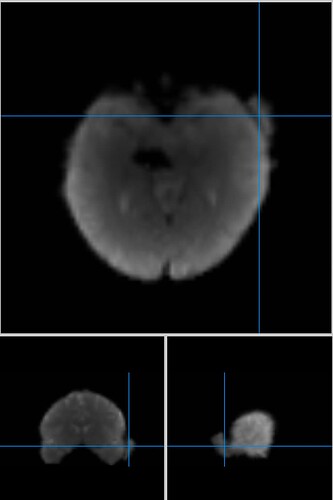
The individual echo images (_echo-*_desc-preproc_bold.nii) aren’t skull-stripped so it doesn’t look like the problem is carried over from fMRIPrep. However, the tedana-derived optimally-combined images (and their masks *desc-adaptiveGoodSignal_mask.nii ) show the same problem.
I have many questions…
1. What might be causing fMRIPrep’s overly-inclusive BOLD masks? Is there a way to correct it (if it matters)? Would it be expected to affect anything later in the workflow, particularly the transform matrices that I use later for normalizing the Tedana denoised output?
2. Is the inclusion of some skull/dura areas in the adaptive mask from Tedana problematic for the ICA-based denoising (because you’re including areas of signal of no interest in the data for decomposition)?
3. My current workflow is Tedana → normalize denoised OC image to MNI space. If I wanted to use the subject’s own mask from fMRIPrep as an explicit mask, would it make sense to reverse that – so normalize the individual echoes first to MNI space, then specify --mask *_MNI152NLin2009cAsym_desc-brain_mask.nii.gz as the mask for tedana? (Or is there a better way to generate a native space mask that doesn’t involve me re-running fMRIPrep on 73+ subjects x 4 tasks w/1-4 runs each? ![]() )
)
4. Alternatively, would you recommend just masking out the extraneous non-brain areas later, during the single-subject-level analysis, using the participant’s normalized T1w mask from /anat/?
I also considered tweaking the compute_epi_mask parameters in tedana, but I’m reluctant to make them more conservative given that the Tedana adaptive mask often has more-than-ideal dropout.
fMRIPrep details:
- fMRIPrep version: 21.0.2
- fMRIPrep command:
/opt/conda/bin/fmriprep /data/BIDS_data /data/BIDS_data/derivatives/fmriprep participant --participant-label 003 --fs-license-file /hpc/users/seeles01/minerva/license.txt --skip_bids_validation --me-output-echos --n_cpus 16 --omp-nthreads 2 --mem-mb 120000 -vv --notrack
Functional data:
- Original orientation: LAS
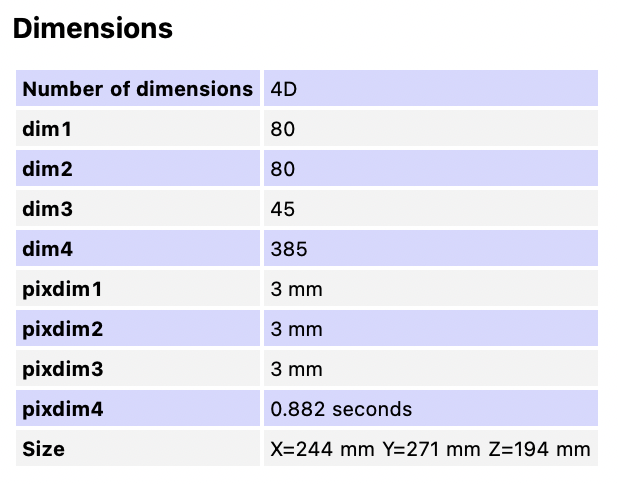
- Repetition time (TR): 0.882s
- Phase-encoding (PE) direction: Anterior-Posterior
- Multi-echo EPI sequence: 4 echoes.
- Slice timing correction: Applied
- Susceptibility distortion correction: PEB/PEPOLAR (phase-encoding based / PE-POLARity)
- Registration: FreeSurfer
bbregister(boundary-based registration, BBR) - 6 dof- Non-steady-state volumes: 2
tedana details:
tedana -d
$sub"_task-"$task"_dir-AP_run-"$r"_echo-1_desc-preproc_bold.nii.gz"
$sub"_task-"$task"_dir-AP_run-"$r"_echo-2_desc-preproc_bold.nii.gz"
$sub"_task-"$task"_dir-AP_run-"$r"_echo-3_desc-preproc_bold.nii.gz"
$sub"_task-"$task"_dir-AP_run-"$r"_echo-4_desc-preproc_bold.nii.gz"
-e 11.0 29.7 48.4 67.1 --tedpca “kundu-stabilize”
–out-dir $sub_outdir
–debug | tee $sub"_task-"$task"_run-"$r"_tedana.log"
Example from fMRIPrep report:
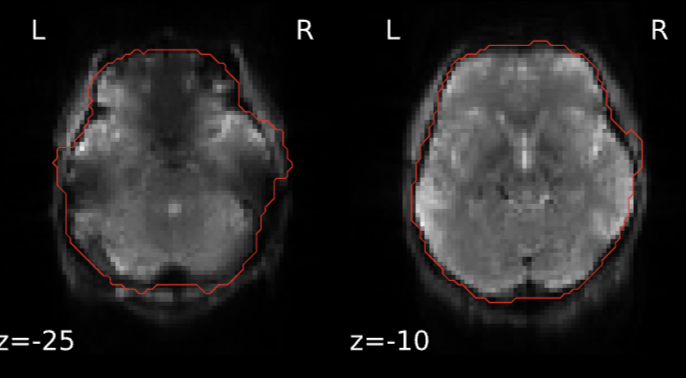
Example: optimally-combined image from Tedana (in native space)