Summary of what happened:
I have been using the recommendations from mumfordbrainstats to adapt fMRIprep-processed data into my FEAT analysis pipeline.
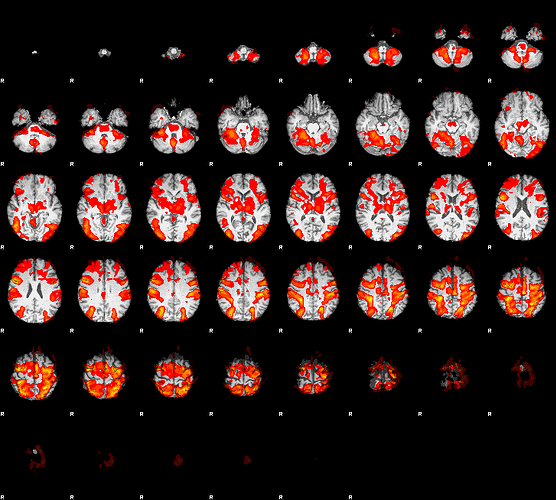
Everything works fine EXCEPT that my higher-level background images, and thus all the .png files, are on a smoothed background. The functional data look fine: voxels sizes are correct, the data passes Mumford’s data checks, and activity map looks very similar to a FSL-only analysis. Therefore I think it’s not affecting my results, just my visualizations; i.e. bg_image is from smoothed data.
Anyone know if there’s a way to avoid this? Or must I just avoid & ignore the automatically-generated backgrounds?
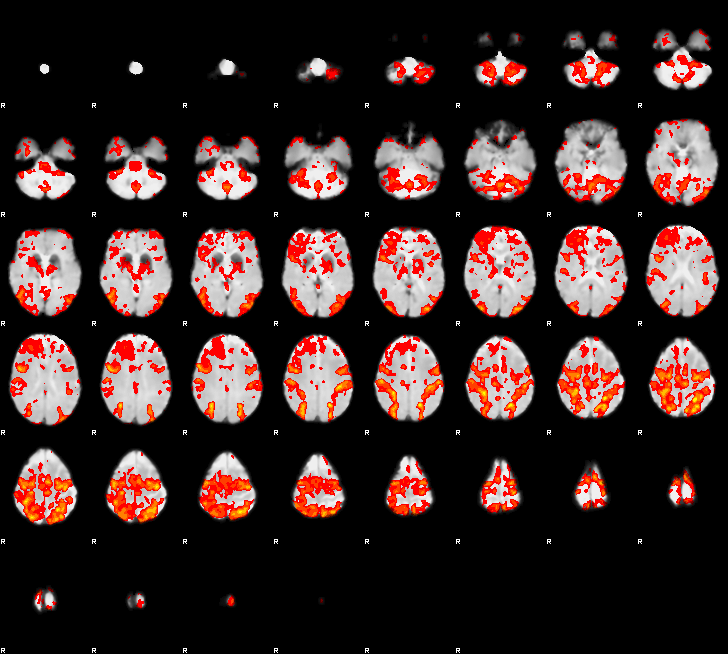
Here’s the rendered_thresh_zstat1 from a FSL-only pipeline, looking as it should:
Here’s the rendered_thresh_zstat1.png from the fMRIprep-to-FSL pipeline. Activity map is very similar (despite some registration differences), but the anatomical background image is smoothed.