Summary of what happened:
Hello!
I’m running into inconsistencies when analyzing fMRIPrep-preprocessed data in FSL. When I preprocessed the same data directly in FSL, the results differ.
How I used the fMRIPrep-preprocessed data in FSL:
- I selected the functional output from fMRIPrep that was already in MNI space
- I then registered this functional output to the preprocessed MNI-space T1-weighted image using FSL
- Using the FEAT directories from the registration output, I ran a first-level analysis, including additional confound EVs: translation (trans_x, trans_y, trans_z), rotation (rot_x, rot_y, rot_z), and nuisance signals (CSF, white matter, global signal)
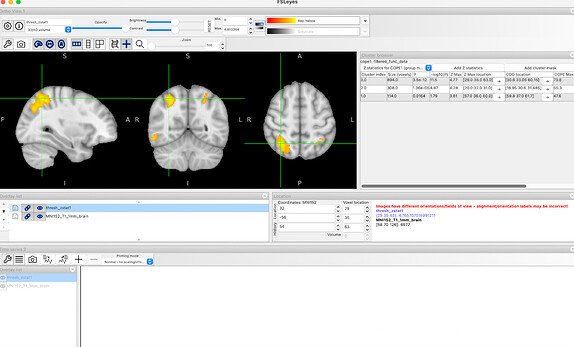
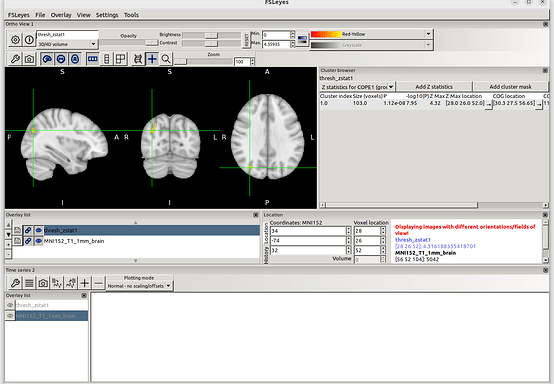
- Using the cope3 outputs from the first-level analysis I ran the third-level analysis (the GLMs were the same for both analyses)
Version:
FSL 6.0.7.14
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS
Screenshots / relevant information:
Results from FSL preprocessing:
Results from fMRIprep preprocessing: