Summary of what happened:
I seem to be running into a problem with fmriprep and (I believe) the fieldmaps that are being applied. I have received the following error:
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node topup.
Cmdline:
topup --config=/opt/conda/lib/python3.9/site-packages/sdcflows/data/flirtsch/b02b0.cnf --datain=/scratch/fmriprep_23_0_wf/single_subject_0005_wf/fmap_preproc_wf/wf_auto_00000/topup/sub-0005_acq-smallFOVcor_dir-FH_epi_average_merged_sliced_volregLAS_encfile.txt --imain=/scratch/fmriprep_23_0_wf/single_subject_0005_wf/fmap_preproc_wf/wf_auto_00000/to_las/sub-0005_acq-smallFOVcor_dir-FH_epi_average_merged_sliced_volregLAS.nii.gz --out=sub-0005_acq-smallFOVcor_dir-FH_epi_average_merged_sliced_volregLAS_base --iout=sub-0005_acq-smallFOVcor_dir-FH_epi_average_merged_sliced_volregLAS_corrected.nii.gz --fout=sub-0005_acq-smallFOVcor_dir-FH_epi_average_merged_sliced_volregLAS_field.nii.gz --jacout=jac --logout=sub-0005_acq-smallFOVcor_dir-FH_epi_average_merged_sliced_volregLAS_topup.log --rbmout=xfm --dfout=warpfield
Stdout:
Stderr:
Error occurred when preparing to run topup
Exception thrown with message: Topup: msg=TopupScan::TopupScan: third element of pevec must be zero
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 453, in aggregate_outputs
setattr(outputs, key, val)
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/traits_extension.py", line 330, in validate
value = super(File, self).validate(objekt, name, value, return_pathlike=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/traits_extension.py", line 135, in validate
self.error(objekt, name, str(value))
File "/opt/conda/lib/python3.9/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: The 'out_movpar' trait of a TOPUPOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '/scratch/fmriprep_23_0_wf/single_subject_0005_wf/fmap_preproc_wf/wf_auto_00000/topup/sub-0005_acq-smallFOVcor_dir-FH_epi_average_merged_sliced_volregLAS_base_movpar.txt' <class 'str'> was specified.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 400, in run
outputs = self.aggregate_outputs(runtime)
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 460, in aggregate_outputs
raise FileNotFoundError(msg)
FileNotFoundError: No such file or directory '/scratch/fmriprep_23_0_wf/single_subject_0005_wf/fmap_preproc_wf/wf_auto_00000/topup/sub-0005_acq-smallFOVcor_dir-FH_epi_average_merged_sliced_volregLAS_base_movpar.txt' for output 'out_movpar' of a TOPUP interface
Command used (and if a helper script was used, a link to the helper script or the command generated):
The command I have input is the following:
fmriprep-docker /lbc/lbc1/derivatives/brainstem_funk/BIDS /lbc/lbc1/derivatives/brainstem_funk/fmriprep participant -w work/ --fs-license-file /lbc/lbc1/derivatives/brainstem_funk/license.txt --fs-no-reconall --skip-bids-validation --output-spaces MNI152NLin2009cAsym:res-native MNI152NLin2009cAsym
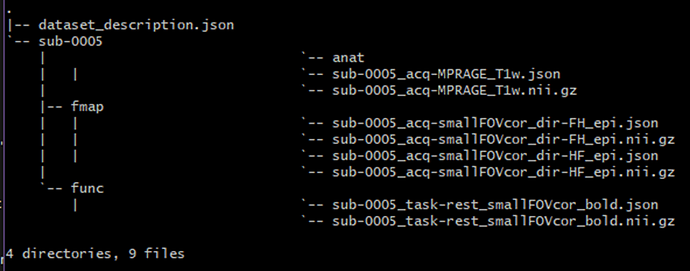
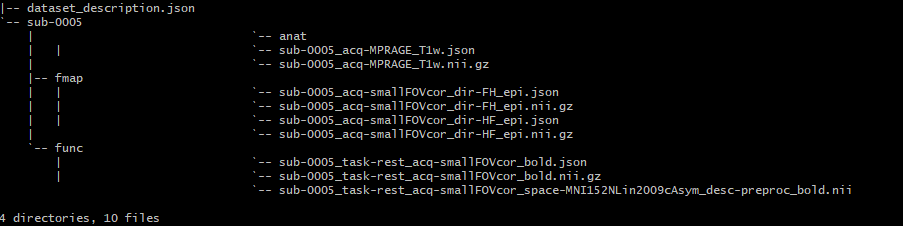
The BIDS folder is set up as follows:
**(I’m new to bash so I don’t know why ‘anat’ looks like a file in the tree; It’s definitely a folder in the BIDS file)
Version:
- fmriprep v. 23.0.2
- docker v. 20.10.21
Environment (Docker, Singularity, custom installation):
Installed using docker.
Data formatted according to a validatable standard? Please provide the output of the validator:
It is listed as valid according to bids.validator.
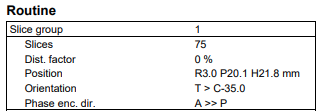
Screenshots / relevant information:
I have included the “IntendedFor” with the file path in the .json files.