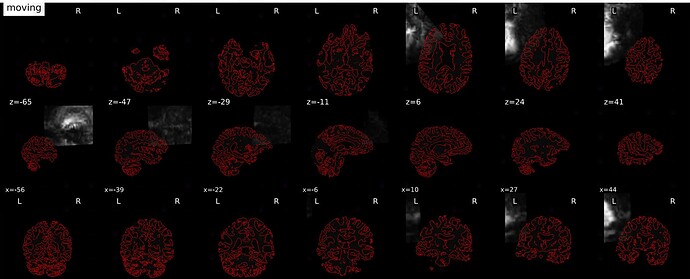
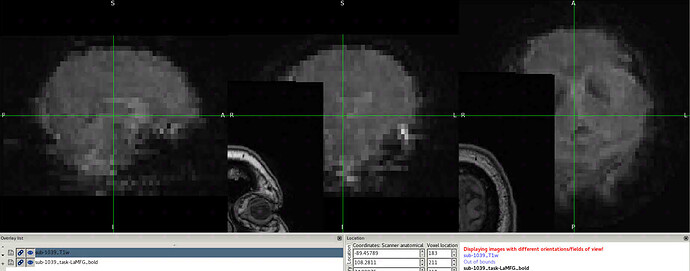
Summary of what happened:
I am running fmriprep jobs for multiple subjects and some subjects get the following error:
traits.trait_errors.TraitError: The 'md1d_file' trait of a VolregOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '/Shared/jianglab/3_Data_Working/CausalConnectome/tmsfmri_fmriprep_test/workdir/fmriprep_22_0_wf/single_subject_3074_wf/func_preproc_task_LpMFG_wf/bold_t1_trans_wf/bold_reference_wf/gen_avg/vol0000_xform-00000_clipped_merged_sliced_md.1D' <class 'str'> was specified.
I checked the input file and confirmed there are no issues with them. There is also a .html output for that subject and it looks correct.
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv \
--bind $bids_root_dir \
/Shared/jianglab/0_scripts/Xin/fmri_prep_guochun/fmriprep-22.0.2.simg \
$bids_root_dir/bidsdata $bids_root_dir/fmriprep \
participant \
--participant-label $subj \
--output-spaces MNI152NLin2009cAsym:res-2 \
--stop-on-first-crash \
--use-plugin $bids_root_dir2/scripts/plugin.yml \
--ignore slicetiming \
--fs-license-file $bids_root_dir2/code/license.txt \
--resource-monitor \
--low-mem \
--force-bbr \
--fs-no-reconall \
-w $bids_root_dir/workdir >
Version:
You are using fMRIPrep-22.0.2, and a newer version of fMRIPrep is available: 23.0.2.
bids-validator@1.8.0
Environment (Docker, Singularity, custom installation):
it runs on a singularity container
Data formatted according to a validatable standard? Please provide the output of the validator:
The input is in BIDS format. Other files with the same format run correctly
Relevant log outputs (up to 20 lines):
raceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 454, in aggregate_outputs
setattr(outputs, key, val)
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/traits_extension.py", line 330, in validate
value = super(File, self).validate(objekt, name, value, return_pathlike=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/traits_extension.py", line 135, in validate
self.error(objekt, name, str(value))
File "/opt/conda/lib/python3.9/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: The 'md1d_file' trait of a VolregOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '/Shared/jianglab/3_Data_Working/CausalConnectome/tmsfmri_fmriprep_test/workdir/fmriprep_22_0_wf/single_subject_3074_wf/func_preproc_task_LpMFG_wf/bold_t1_trans_wf/bold_reference_wf/gen_avg/vol0000_xform-00000_clipped_merged_sliced_md.1D' <class 'str'> was specified.