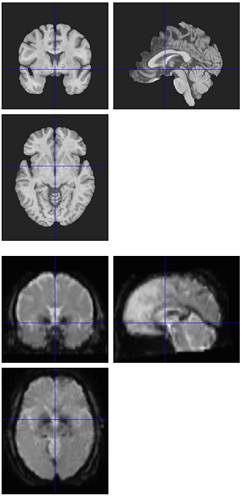
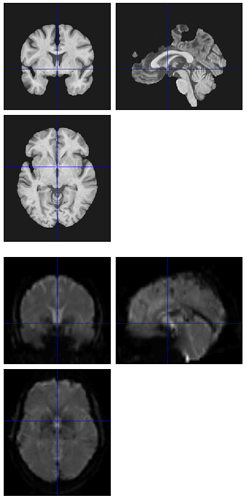
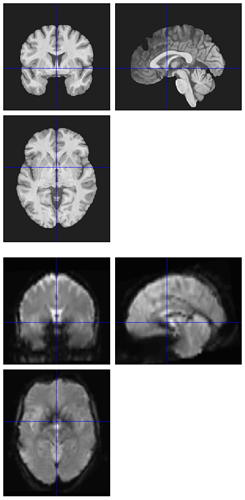
Hi all,
I’m seeing the same issue in quite a few of my fmriprep (1.1.8) reports, where the BBR looks like it ran into trouble:
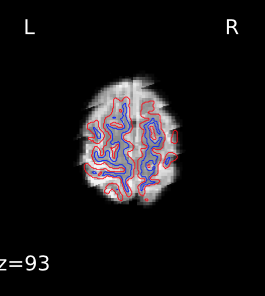
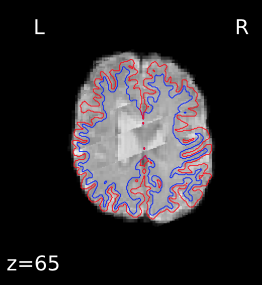
Registration does look okay if I use --fs-no-reconall for the affected participants, at least the ones I’ve done so far. Example .svg files and report here: https://www.dropbox.com/sh/5q20redzlltxm9j/AABRXhWUBxBHxWJJPLcZaSEfa?dl=0)
These are the commands used for preprocessing:
With Freesurfer bbr:
/usr/local/miniconda/bin/fmriprep /data /out participant --low-mem --resource-monitor --stop-on-first-crash --longitudinal --use-syn-sdc --use-aroma --participant_label sub-122 sub-123 sub-125 -w /scratch
No Freesurfer (FLIRT):
/usr/local/miniconda/bin/fmriprep /data /out participant --low-mem --resource-monitor --stop-on-first-crash --fs-no-reconall --longitudinal --use-syn-sdc --use-aroma --participant_label sub-122 sub-123 sub-125 -w /scratch
Is this an issue with the visualization only, or does it indicate an actual issue with the registration? It’s interesting that for the affected participants, I see the same problem in the same areas and in approximately the same slices. It doesn’t seem to be particularly related to how much movement there is in the BOLD series. I also don’t see a clear pattern of affected subjects being outliers or extreme values on the MRIQC group report metrics. Some of the affected people do have lower CNR in their structural images, but at least one person has fairly high CNR (relative to the group). The input images don’t appear to be especially distorted.
Any ideas what might be causing this, and/or recommendations to address? (I’d like to continue using Freesurfer BBR because it was working much better for a lot of these older adult brains with structural differences from the T1 template that FLIRT was having trouble coping with. Unless it’s okay to use Freesurfer for some and not for others, depending on which yields the best registration – but that seems a bit suspect to me, to use different pipelines within the same dataset.)
Thanks for any insight you can offer!