Hi everyone,
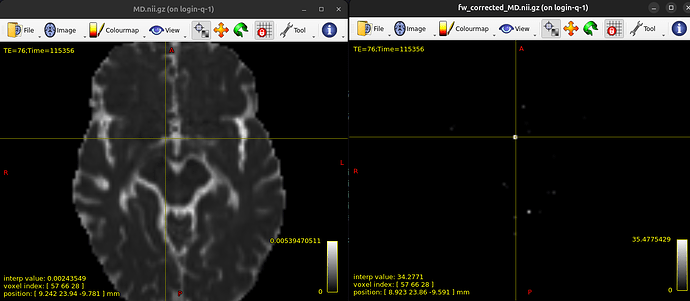
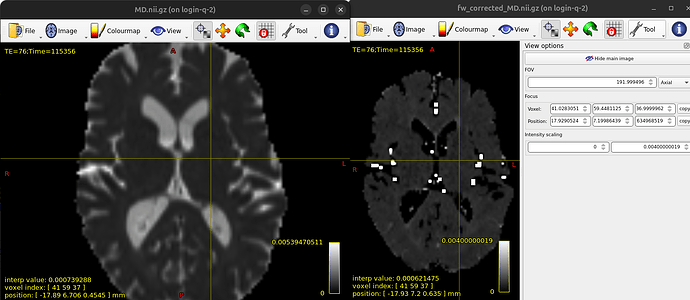
I ran fwdtifit in DIPY and the results for FA look good but the MD images turn out extremely dark. When I compare them to the images I get with dtifit, it seems like the brightest voxels in the free water corrected MD image correspond to bright voxels in the original MD image and it seems like a scale issue because the intensities for bright voxels are extremely high while the intensities for most voxels are normal. I’m not sure if something went wrong and would appreciate ideas!
I used b-values of 0, 300 and 1000 and the code that is in the documentation
https://docs.dipy.org/stable/examples_built/reconstruction/reconst_fwdti.html#sphx-glr-examples-built-reconstruction-reconst-fwdti-py
EDIT:
I just realised that if I rescale the intensities (between 0 and 0.004 seems to work well) the FW corrected MD image looks almost as expected except for the few very bright bits. In the documentation it says “Unexpected high amplitudes of FA are however observed in the periventricular gray matter. This is a known artefact of regions associated to voxels with high water volume fraction (i.e. voxels containing basically CSF).” - could it be that something similar is happening in my MD image too and that I can just remove these voxels in the same way?