Hi everyone,
I am trying to convert some fMRI data in the CVS (cvs_avg35) space to the MNI125 standard. This is so that I could compare the data currently in CVS to other datasets in MNI space. I have made an attempt to register CVS data onto MNI space and the output appears to look a little better but I’m not sure if I followed the correct registration process or if my output needs further improvement. Any advice is appreciated!
CVS data source website: Group-Constrained Subject-Specific (GSS) parcels
CVS data download: Object parcels
Steps I followed using Freesurfer:
Registering cvs_avg35 template to mni125 space:
mni152reg --s cvs_avg35
This generated reg.mni152.2mm.dat in
/freesurfer/subjects/cvs_avg35/mri/transforms/Registering CVS subject data to MNI space using the .dat file
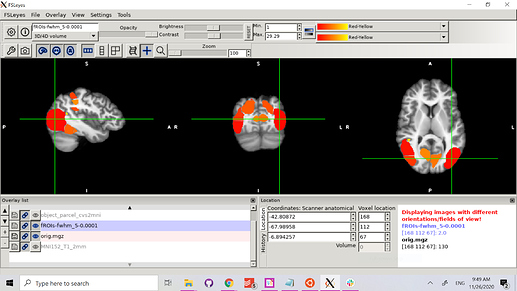
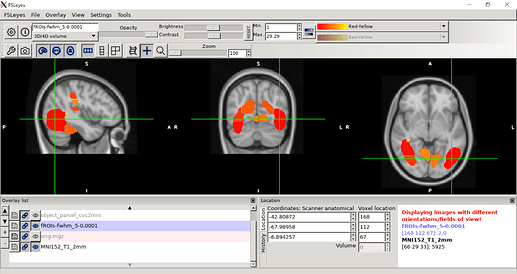
mri_vol2vol --mov $SUBJECTS_DIR/fROIs-fwhm_5-0.0001.nii --o $OUTPUT_DIR/object_parcel_cvs2mni.nii --reg $FREESURFER_HOME/subjects/cvs_avg35/mri/transforms/reg.mni152.2mm.dat --mni152reg
When viewing the output file overlayed on an MNI template in fsleyes, the result looked inverted as all the orientations seemed flipped. I had to redo this step another way by inverting the registration source and target.
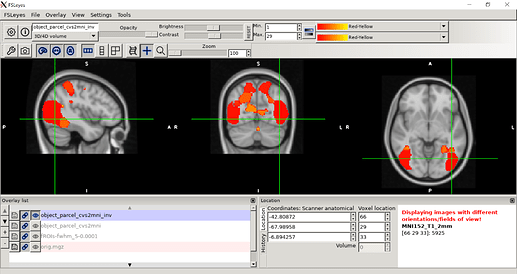
(Inverse) Registering CVS subject data to MNI space using the .dat file
mri_vol2vol --targ $SUBJECTS_DIR/fROIs-fwhm_5-0.0001.nii --mov $FSLDIR/data/standard/MNI152_T1_2mm.nii.gz --o
$OUTPUT_DIR/object_parcel_cvs2mni_inv.nii --inv --reg $FREESURFER_HOME/subjects/cvs_avg35/mri/transforms/reg.mni152.2mm.dat
Here’s what the CVS subject data looks like on a cvs_avg35 template
Here’s what the CVS subject data looks like on a mni125 template
Finally, CVS subject data after registering to MNI space (in step 3)