Summary of what happened:
A freesurfer error is causing fmriprep to fail. The rh and the left half errors slightly differently, but both seem related.
LH error:
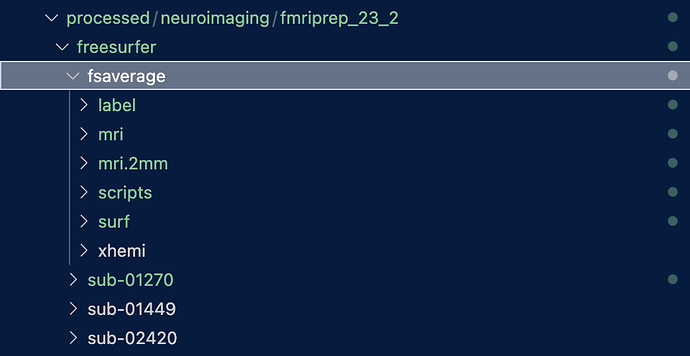
ERROR: Label BA1_exvivo does not exist in SUBJECTS_DIR fsaverage! The fsaverage link probably points to an older freesurfer version
RH error:
error: mri_label2label: could not open label file /out/freesurfer/fsaverage/label/rh.BA2_exvivo.thresh.label ERROR reading /out/freesurfer/fsaverage/label/rh.BA2_exvivo.thresh.label Linux qnode0402 3.10.0-1160.95.1.el7.x86_64 #1 SMP Fri Jun 23 08:44:55 EDT 2023 x86_64 x86_64 x86_64 GNU/Linux
Command used (and if a helper script was used, a link to the helper script or the command generated):’
list_subs.txt is a text file container a list of subjects.
#!/usr/bin/bash
#SBATCH -A p99999
#SBATCH -p normal
#SBATCH -t 16:00:00
#SBATCH --array=0-4%10
#SBATCH --job-name="fmriprep_TEAM_\${SLURM_ARRAY_TASK_ID}"
#SBATCH --output=fmriprep.%A_%a.out
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=4
#SBATCH --mem=15G
module purge
module load singularity/latest
IFS=$'\n' read -d '' -r -a input_args < list_subs.txt
#WITHOUT FMAP
singularity run --cleanenv -B /projects/b1108:/projects/b1108 \
-B /projects/b1108/studies/TEAM/data/processed/neuroimaging/fmriprep_23_2:/out \
-B /projects/b1108/studies/TEAM/data/raw/neuroimaging/bids:/data \
-B /projects/b1108/studies/TEAM/data/processed/neuroimaging/fmriprep_23_2/work:/work \
/projects/b1108/software/singularity_images/fmriprep_23.2.0.sif \
/data /out participant --nthreads 4 --omp-nthreads 3 --mem_mb 30000 \
--participant-label ${input_args[$SLURM_ARRAY_TASK_ID]} \
--fs-license-file /projects/b1108/software/freesurfer_license/license.txt \
--output-spaces MNI152NLin6Asym \
--fs-subjects-dir /out/freesurfer \
-w /work --ignore fieldmaps
Version:
23.2.0 for fmriprep
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
bids-validator@1.9.3
bids-specification@disable
e[36m Please visit https://neurostars.org/search?q=INCONSISTENT_SUBJECTS for existing conversations about this issue.e[39m
e[33m3: [WARN] Not all subjects/sessions/runs have the same scanning parameters. (code: 39 - INCONSISTENT_PARAMETERS)e[39m
./sub-09001/ses-1/func/sub-09001_ses-1_task-mid_run-1_bold.nii.gz
The most common set of dimensions is: 64,64,35,292 (voxels), This file has the dimensions: 64,64,35,368 (voxels).
./sub-09002/ses-1/func/sub-09002_ses-1_task-mid_run-1_bold.nii.gz
The most common set of dimensions is: 64,64,35,292 (voxels), This file has the dimensions: 64,64,35,368 (voxels).
./sub-09993/ses-1/func/sub-09993_ses-1_task-mid_run-1_bold.nii.gz
The most common set of dimensions is: 64,64,35,292 (voxels), This file has the dimensions: 64,64,35,368 (voxels).
./sub-09993/ses-1/func/sub-09993_ses-1_task-rest_bold.nii.gz
The most common set of dimensions is: 64,64,35,300 (voxels), This file has the dimensions: 64,64,35,247 (voxels).
./sub-15305/ses-1/func/sub-15305_ses-1_task-mid_run-1_bold.nii.gz
The most common set of dimensions is: 64,64,35,292 (voxels), This file has the dimensions: 64,64,35,43 (voxels).
./sub-15416/ses-1/func/sub-15416_ses-1_task-mid_run-1_bold.nii.gz
The most common set of dimensions is: 64,64,35,292 (voxels), This file has the dimensions: 64,64,35,368 (voxels).
./sub-15427/ses-1/func/sub-15427_ses-1_task-mid_run-1_bold.nii.gz
The most common set of dimensions is: 64,64,35,292 (voxels), This file has the dimensions: 64,64,35,547 (voxels).
./sub-15658/ses-1/fmap/sub-15658_ses-1_magnitude1.nii.gz
The most common set of dimensions is: 78,78,35 (voxels), This file has the dimensions: 78,78,39 (voxels).
./sub-15658/ses-1/fmap/sub-15658_ses-1_magnitude2.nii.gz
The most common set of dimensions is: 78,78,35 (voxels), This file has the dimensions: 78,78,39 (voxels).
./sub-15658/ses-1/fmap/sub-15658_ses-1_phasediff.nii.gz
The most common set of dimensions is: 78,78,35 (voxels), This file has the dimensions: 78,78,39 (voxels).
./sub-15658/ses-1/func/sub-15658_ses-1_task-mid_run-1_bold.nii.gz
The most common set of dimensions is: 64,64,35,292 (voxels), This file has the dimensions: 64,64,34,292 (voxels).
./sub-15909/ses-1/func/sub-15909_ses-1_task-mid_run-2_bold.nii.gz
The most common set of dimensions is: 64,64,35,292 (voxels), This file has the dimensions: 64,64,35,259 (voxels).
./sub-18215/ses-1/anat/sub-18215_ses-1_T1w.nii.gz
The most common set of dimensions is: 176,512,512 (voxels), This file has the dimensions: 160,512,512 (voxels).
e[36m Please visit https://neurostars.org/search?q=INCONSISTENT_PARAMETERS for existing conversations about this issue.e[39m
e[33m4: [WARN] The recommended file /README is missing. See Section 03 (Modality agnostic files) of the BIDS specification. (code: 101 - README_FILE_MISSING)e[39m
e[36m Please visit https://neurostars.org/search?q=README_FILE_MISSING for existing conversations about this issue.e[39m
e[34me[4mSummary:e[24me[39m e[34me[4mAvailable Tasks:e[24me[39m e[34me[4mAvailable Modalities:e[24me[39m
2136 Files, 26.95GB gng MRI
137 - Subjects mid
1 - Session rest
e[36m If you have any questions, please post on https://neurostars.org/tags/bids.e[39m
Relevant log outputs (up to 20 lines):
LH log output:
9315 6354 20345 2.783 0.693 0.106 0.020 78 8.0 superiortemporal
6100 4309 11768 2.577 0.467 0.117 0.022 57 5.7 supramarginal
855 537 1335 2.285 0.384 0.119 0.023 9 0.7 transversetemporal
3204 2135 6764 3.133 0.724 0.106 0.023 25 2.9 insula
/out/freesurfer/sub-01270/label
#--------------------------------------------
#@# BA_exvivo Labels lh Wed May 1 14:42:01 CDT 2024
-------------------------------------------------------------------------
ERROR: Label BA1_exvivo does not exist in SUBJECTS_DIR fsaverage!
The fsaverage link probably points to an older freesurfer version
-------------------------------------------------------------------------
Linux qnode0402 3.10.0-1160.95.1.el7.x86_64 #1 SMP Fri Jun 23 08:44:55 EDT 2023 x86_64 x86_64 x86_64 GNU/Linux
recon-all -s sub-01270 exited with ERRORS at Wed May 1 14:42:01 CDT 2024
To report a problem, see http://surfer.nmr.mgh.harvard.edu/fswiki/BugReporting
RH Log output:
mri_label2label --srcsubject fsaverage --srclabel /out/freesurfer/fsaverage/label/rh.BA2_exvivo.thresh.label --trgsubject sub-01270 --trglabel ./rh.BA2_exvivo.thresh.label --hemi rh --regmethod surface
No such file or directory
srclabel = /out/freesurfer/fsaverage/label/rh.BA2_exvivo.thresh.label
srcsubject = fsaverage
trgsubject = sub-01270
trglabel = ./rh.BA2_exvivo.thresh.label
regmethod = surface
srchemi = rh
trghemi = rh
trgsurface = white
srcsurfreg = sphere.reg
trgsurfreg = sphere.reg
usehash = 1
Use ProjAbs = 0, 0
Use ProjFrac = 0, 0
DoPaint 0
SUBJECTS_DIR /out/freesurfer
FREESURFER_HOME /opt/freesurfer
Loading source label.
error: Invalid argument
error: mri_label2label: could not open label file /out/freesurfer/fsaverage/label/rh.BA2_exvivo.thresh.label
ERROR reading /out/freesurfer/fsaverage/label/rh.BA2_exvivo.thresh.label
Linux qnode0402 3.10.0-1160.95.1.el7.x86_64 #1 SMP Fri Jun 23 08:44:55 EDT 2023 x86_64 x86_64 x86_64 GNU/Linux
recon-all -s sub-01270 exited with ERRORS at Wed May 1 14:46:21 CDT 2024
To report a problem, see http://surfer.nmr.mgh.harvard.edu/fswiki/BugReporting
Screenshots / relevant information:
Looks like I’m experiencing the same issue as these folks: ERROR: Label BA1_exvivo does not exist in SUBJECTS_DIR fsaverage! The fsaverage link probably points to an older freesurfer version · Issue #3258 · nipreps/fmriprep · GitHub. I added --fs-subjects-dir /output/freesurfer to my fmriprep call and still encountered the same error. I have tested with multiple participants.