Hello,
I am seeking guidance on the best practices for using FSL’s Brain Extraction Tool (BET) for skull stripping on T2-weighted brain images that contain a Glioblastoma (GBM) tumor.
My objective is to create an accurate brain mask that includes the entire brain parenchyma, the tumor, and any surrounding edema, while excluding the skull and non-brain tissue.
Software and System Details:
FSL Version: [6.0.7.18]
Operating System: Ubuntu [22.04]
Image Type: T2-weighted NIfTI (.nii)
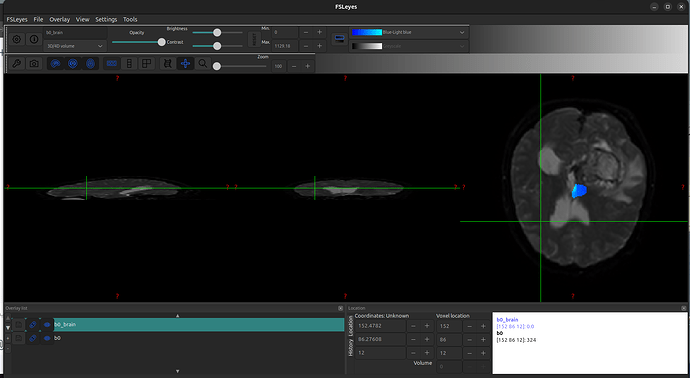
The Problem: When I run the standard BET command, the resulting brain mask is too aggressive. It incorrectly removes parts of the brain, especially in the area of the tumor and the surrounding bright edema. The mask essentially “cuts into” the tissue I need to keep.
What I Have Tried So Far:
- I have run the standard
betcommand:bet input_T2_image.nii.gz output_brain.nii.gz - I have tried adjusting the fractional intensity threshold (
-fparameter), but I haven’t found a value that works well. A low value includes the skull, and a high value removes even more brain tissue. For example:bet input_T2_image.nii.gz output_brain_f0.3.nii.gz -f 0.3
My Question: What is the recommended approach or set of parameters for using BET on T2 images with hyperintense lesions like GBM? Are there alternative options within the FSL suite (e.g., BET with the -R robust option, or another tool entirely) that are better suited for this challenging task?
Thank you for any advice you can provide.
Here is a screenshot showing the problem: