Hi,
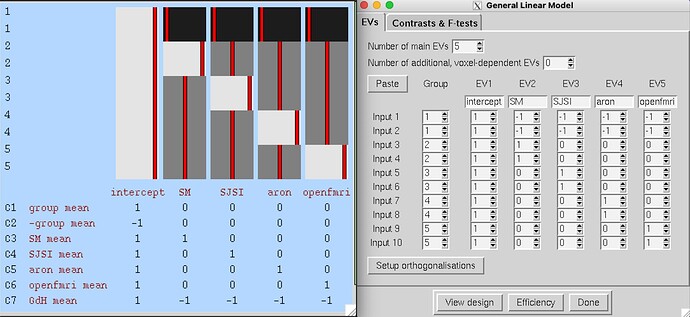
I am trying to compute whole brain GLMs using FSL FEAT for data from five different datasets. Each dataset contains stop signal data. I want to build a model that calculates the mean of all datasets together as well as each dataset independently, so that I can compare them to the overall mean and also have a singular SPM that includes data from all datasets. To do this, I have setup dummy variables using a factor effects model based on information I found here: GLM - FslWiki. The dummy variables work as expected, and gives results similar to running each dataset independently as well as giving an overall mean. The problem I have is that when I try to setup different variances groups for each dataset, I get an error saying the design matrix is non-separable.
Error message:
‘Problem with processing the model: Warning - design matrix uses different groups (for different variances), but these do not contain “separable” EVs for the different groups (it is necessary that, for each EV, only one of the groups has non-zero values).’
I was wondering if anyone could advise a way to build this model so that I can include different variance groups for each dataset, calculate a grand mean (intercept) of all datasets, and use these dummy variables so I can estimate intercepts for each dataset independently as well.
I am running FEAT using nipype but I have attached images of the full model setup from the FEAT GUI to make visualising the problem easier.
Let me know if I can provide any additional information.
Best,
Scott