Summary of what happened:
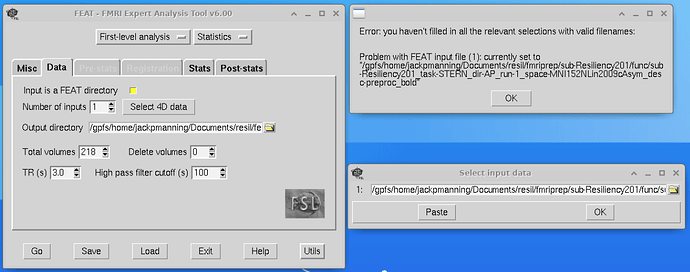
I am trying to load data into FEAT, but it keeps dropping the file extension from the data, and then telling me that I haven’t filled in all the relevant sections with valid filenames.
I am running FSL/FEAT through a virtual machine that is running on Ubuntu 20.04.6 (Focal Fossa), and I am running FEAT v6.00. I cannot alter the FSL or FEAT installations to change versions.
Please help, I am not sure why this is happening, and I haven’t been able to find any answers on here or google.
Screenshots / relevant information:
Hi @jackpmanning dropping the file extension is normal behaviour for many FSL tools, as it will read NIfTI images in a range of different forms (.nii, .nii.gz, .img/.hdr pairs). This is a throwback to the days of ANALYZE.
What is the actual name (with the file extension) of your input file here?
Howdy,
This is the file path
/gpfs/home/jackpmanning/Documents/resil/fmriprep/sub-Resiliency201/func/sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
I am not sure if this matters but I preprocessed the data with fmriprep, and tried to follow mumforbrainstat’s video on how to use fmriprep data in feat (https://www.youtube.com/watch?v=U3tG7JMEf7M&t=310s&ab_channel=mumfordbrainstats)
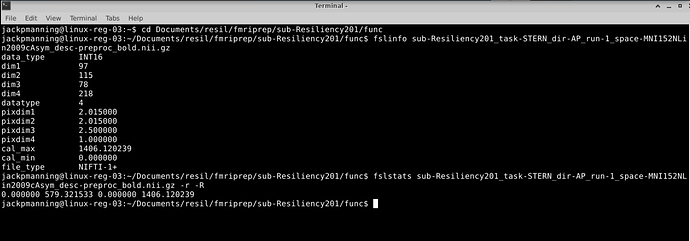
I don’t think there is anything particular about the file name that would cause FEAT to fail. What are the properties of the file - can you run these commands and paste the output?
fslinfo /gpfs/home/jackpmanning/Documents/resil/fmriprep/sub-Resiliency201/func/sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
fslstats /gpfs/home/jackpmanning/Documents/resil/fmriprep/sub-Resiliency201/func/sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz -r -R
Of course, here is the text output:
jackpmanning@linux-reg-03:~ cd Documents/resil/fmriprep/sub-Resiliency201/func
jackpmanning@linux-reg-03:~/Documents/resil/fmriprep/sub-Resiliency201/func$ fslinfo sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
data_type INT16
dim1 97
dim2 115
dim3 78
dim4 218
datatype 4
pixdim1 2.015000
pixdim2 2.015000
pixdim3 2.500000
pixdim4 1.000000
cal_max 1406.120239
cal_min 0.000000
file_type NIFTI-1+
jackpmanning@linux-reg-03:~/Documents/resil/fmriprep/sub-Resiliency201/func$ fslstats sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz -r -R
0.000000 579.321533 0.000000 1406.120239
jackpmanning@linux-reg-03:~/Documents/resil/fmriprep/sub-Resiliency201/func$
and a screenshot of the same thing
And what is the output of pwd from within that same directory? It looks like this is a network mount? Perhaps the path needs to be different in order for it to be correclty interpreted
Another command to run would be:
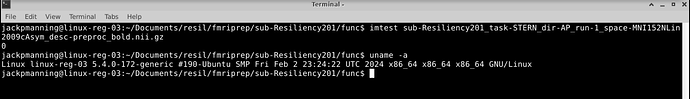
imtest /gpfs/home/jackpmanning/Documents/resil/fmriprep/sub-Resiliency201/func/sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
as the FEAT GUI uses imtest to determine whether the file path is valid. Perhaps there is an issue with the imtest command on your system.
The result of imtest is 0.
I’m pretty sure this is a virtual machine with a network mount (click Texas ViDaL for more info).
Here is some of the system info too.
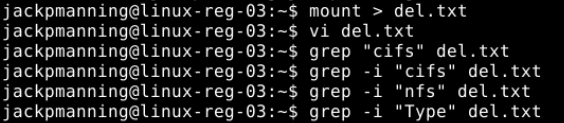
I am not sure how to check if I am on a network mount. I put the contents of ‘mount’ into a .txt file and searched it for “cifs” and “nfs” and they are not present (indicating maybe it is not a network mount?).
The imtest command is returning 0 which means it thinks that the file is not an image. What version of FSL are you using? You can find out by running cat $FSLDIR/etc/fslversion
jackpmanning@linux-reg-03:~$ cat $FSLDIR/etc/fslversion
6.0.7.7
fsleyes will not open the files either. I was able to open the file in mricron, though this was on my local computer, not the virtual machine (it doesn’t have mricron).
Could this be happening because of something fmriprep did? I am quite confused.
edit: I downloaded mricron on the virtual machine and I was able to open the image in it.
What happens when you try to open the image with FSLeyes?
What is the output of running this command:
file /gpfs/home/jackpmanning/Documents/resil/fmriprep/sub-Resiliency201/func/sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
I can open the .nii.gz files in fsleyes no problem (when I tried last time I was using open directory and not open file).
This is the output from the command:
jackpmanning@linux-reg-03:~$ file /gpfs/home/jackpmanning/Documents/resil/fmriprep/sub-Resiliency201/func/sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
/gpfs/home/jackpmanning/Documents/resil/fmriprep/sub-Resiliency201/func/sub-Resiliency201_task-STERN_dir-AP_run-1_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz: gzip compressed data, max compression, original size modulo 2^32 379359592 gzip compressed data, reserved method, has CRC, extra field, has comment, encrypted, from FAT filesystem (MS-DOS, OS/2, NT), original size modulo 2^32 379359592
If there are multiple files with the same name after the file extension is dropped, would I get this error? The outputs of fmriprep give me a .nii.gz and a .nii file for each scan with the same name. When the extension is dropped, there are effectively two files with the same name in the directory. Could this be the root of the issue?
Aah - yes, that is almost certainly the cause - several FSL tools will give up if there is any ambiguity in the file path after the extension is removed. I’m not particularly familiar with fmriprep, so I’m not sure why it would produce both compressed and uncompressed versions of the file. But you should be able to get FEAT working by removing one of them (FSL defaults to using and generating .nii.gz, but you can change the default by setting the FSLOUTPUTTYPE environment variable as outlined at Document)
I will try that next. I also used the MNI152NLin2009cAsym space by default. Should I instead be using the MNI152NLin6Asym space, and could this alternatively be the root of the issue?
If you are only using FEAT for the model fitting (and disabling registration within FEAT), then your choice of standard template should not have any bearing on the behaviour of FEAT.