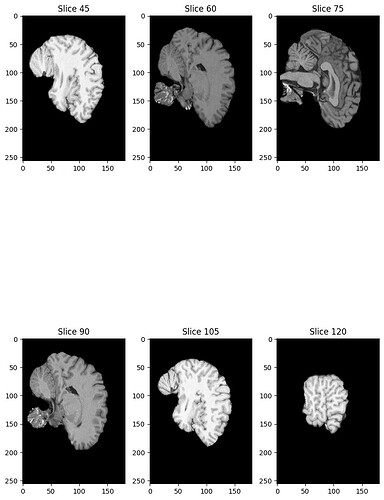
Summary of what happened:
In an MRI (IXI healthy data) preprocessing pipeline containing fslreorient2std and robustfov, I obtained the proper orientation and zoom before applying flirt. However, flirt (post skull-strip) then rotates and zooms in the MRIs. The issue does not apply to all MRIs in the IXI dataset, and I know that the pipeline itself should be fine, since it works perfectly on ADNI data.
IXI dataset is collected in three hospital/institution, Guys, HH and IOP. Three different machines are used. MRIs coming from Guys don’t typically have the above described issue, though a few does. I do encounter most of these issues in HH and IOP MRIs.
Command used:
os.system(f'fslreorient2std {image_path} {preprocessed_image_path}')
os.system(f'robustfov -i {preprocessed_image_path} -r {preprocessed_image_path}')
os.system(f'bet {preprocessed_image_path} {preprocessed_image_path} -R -f {frac_intensity_thres}')
segmented_path = f"{preprocessed_image_path.replace('.nii.gz', '_segout')}"
os.system(f'fast -B -t 1 -o {segmented_path} {preprocessed_image_path}')
preprocessed_image_path = segmented_path + "_restore.nii.gz"
ref = Path(r"icbm_avg_152_t1_tal_lin.nii")
os.system(f'flirt -in {preprocessed_image_path} -ref {ref} -out {preprocessed_image_path}')
frac_intensity_thres was set to 0.5, but I have tried more aggressive skull-stripping, and the problem persists.
Version: Flirt 6.0
Environment: Python making Bash calls
Screenshots / relevant information:
Sagittal slice of an MRI, preprocessed with the pipeline above, post flirt. As a new user I can only upload one image. Between every pre-processing step, I plot a sagittal slice of the brain, thus the above is supposed to be a sagittal slice. The last slice before flirt looks absolutely fine.
Hi @mathinbio, can you share the output of fslhd on both preprocessed_image_path and ref, just before your call to flirt?
Template:
sizeof_hdr 348
data_type FLOAT32
dim0 3
dim1 181
dim2 217
dim3 181
dim4 0
dim5 0
dim6 0
dim7 0
vox_units Unknown
time_units Unknown
datatype 16
nbyper 4
bitpix 32
pixdim0 0.000000
pixdim1 1.000000
pixdim2 1.000000
pixdim3 1.000000
pixdim4 0.000000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 0.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Unknown
qform_code 0
qto_xyz:1 1.000000 0.000000 0.000000 0.000000
qto_xyz:2 0.000000 1.000000 0.000000 0.000000
qto_xyz:3 0.000000 0.000000 1.000000 0.000000
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Left-to-Right
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Unknown
sform_code 3
sto_xyz:1 1.000000 0.000000 0.000000 -90.000000
sto_xyz:2 0.000000 1.000000 0.000000 -126.000000
sto_xyz:3 0.000000 0.000000 1.000000 -72.000000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Left-to-Right
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip mnc2nii icbm_avg_152_t1_tal_lin.mnc icbm_avg_152_t1_tal_lin.nii
aux_file
Before calling flirt:
sizeof_hdr 348
data_type FLOAT32
dim0 3
dim1 146
dim2 256
dim3 181
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 16
nbyper 4
bitpix 32
pixdim0 -1.000000
pixdim1 1.200000
pixdim2 0.937500
pixdim3 0.937509
pixdim4 0.000000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -1.198232 -0.024634 -0.044517 94.909119
qto_xyz:2 -0.031496 0.937177 -0.001172 -87.795753
qto_xyz:3 -0.057001 0.000001 0.936451 -63.057995
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -1.198231 -0.024634 -0.044519 94.908936
sto_xyz:2 -0.031496 0.937177 -0.001173 -87.795868
sto_xyz:3 -0.057004 0.000000 0.936451 -63.057636
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip 2203.12-dirty 2024-02-01T16:17:47+00:00
aux_file
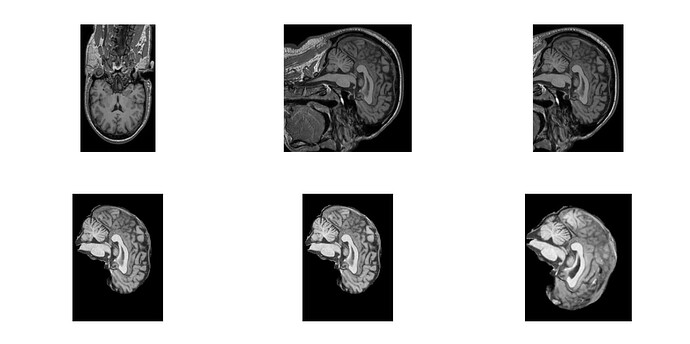
Hi @mathinbio, that looks reasonable. Can you share some screenshots of preprocessed_image_path just before calling flirt? Something like this would be useful:
fsleyes render -of screenshot.png -slightbox -zr 0 1 -sz 1500 1500 <imagefile>
I have an error carrying out the above command, something about OSMesa. Anyway will the ones below work?
Before flirt:
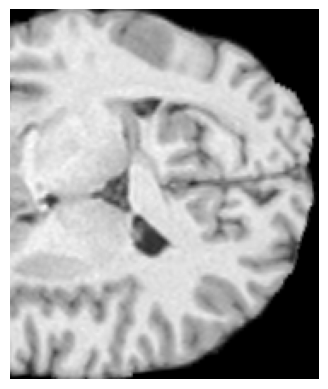
It looks like brain extraction hasn’t performed well for this image - there is a good chance that this is what is causing the flirt failure. You may need to adjust the bet parameters on a per-subject basis, or use a different tool for brain extraction (e.g. SynthStrip seems to be a popular choice these days).
Hi @paulmccarthy, it works well for the few images I tried it on, thanks! Will run it on the whole dataset and will give an update.
Hi @paulmccarthy, unfortunately still for some images this happens:
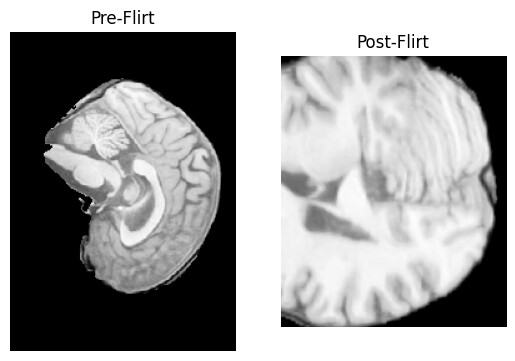
I also upload the header of said image pre flirt.
sizeof_hdr 348
data_type FLOAT32
dim0 3
dim1 146
dim2 256
dim3 181
dim4 1
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 16
nbyper 4
bitpix 32
pixdim0 -1.000000
pixdim1 1.200000
pixdim2 0.937504
pixdim3 0.937501
pixdim4 0.000000
pixdim5 1.000000
pixdim6 1.000000
pixdim7 1.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 0
freq_dim 0
slice_dim 0
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -1.199408 0.027438 -0.010734 85.375122
qto_xyz:2 0.035118 0.937103 0.000315 -96.194366
qto_xyz:3 -0.013746 -0.000000 0.937439 -56.546322
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -1.199408 0.027438 -0.010729 85.375122
sto_xyz:2 0.035118 0.937103 0.000315 -96.194366
sto_xyz:3 -0.013739 0.000000 0.937439 -56.546322
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip 2203.12-dirty 2024-02-01T16:17:47+00:00
aux_file
This is the MNI152 template I’m using. Potentially the problem?
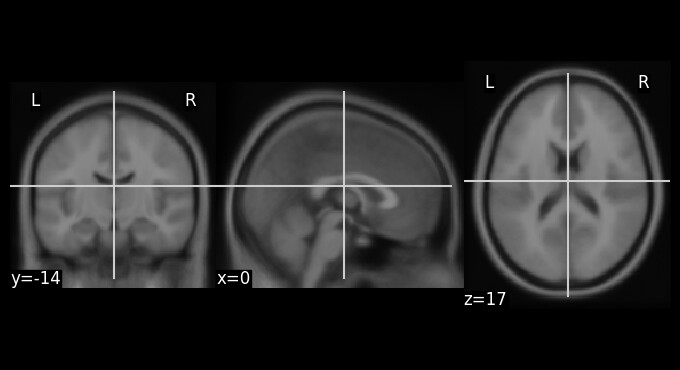
Would you be able to share one of your problematic images, along with the MNI152 template?