Hey all,
I’m currently developing a pipeline to replicate the Heilbron and al study. (10.1073/pnas.2201968119).
I would like to use MNE-python instead of fieldtrip to run my source reconstruction pipeline. However, authors used a specific atlas: atlas_subparc374.R.32k_fs_LR.label.gii available here: derivatives/atlas/atlas_subparc374.R.4k_fs_LR.label.gii
As MNE-python uses Freesurfer files for its source space analyses, I would like to transform this atlas in my freesurfer subject space (.annot or .label files). After several tries, I didn’t managed to achieve this goal.
As far as I understand, fs_LR is the template of the connectome workbench tool, and I was able to get additional files from this template, including L.sphere.32k_fs_LR.surf.gii ( here )
I tried using multiple combinations of freesurfer and connectome_workbench command line tools to transform the .label.gii file to a freesurfer subject .annot or .label file.
Summary of what happened:
Command used (and if a helper script was used, a link to the helper script or the command generated):
from nipype.interfaces.freesurfer import SurfaceTransform, MRIsConvert
subject_dir = '/data/vf/freesurfer'
fs_LR_labels = fr"/data/Heilbron2022/derivatives/atlas/atlas_subparc374.R.32k_fs_LR.label.gii"
fs_LR_sphere = fr"/data/Heilbron2022_derivatives/workbench-anatomy/R.sphere.32k_fs_LR.surf.gii"
fsaverage_sphere = fr"/data/vf/freesurfer/fsaverage/surf/rh.sphere"
fsaverage_white = fr"/data/vf/freesurfer/fsaverage/surf/rh.white"
subject = "sub-VF"
subject_sphere = fr"/data/vf/freesurfer/sub-VF/surf/rh.sphere"
subject_white = fr"/data/vf/freesurfer/sub-VF/surf/rh.white"
# fsaverage sphere to .gii
convert_sphere = MRIsConvert()
convert_sphere.inputs.in_file = fsaverage_sphere
convert_sphere.inputs.out_file = "rh.sphere.surf.gii"
convert_sphere_results = convert_sphere.run()
fsaverage_sphere_gii = convert_sphere_results.outputs.get()['converted']
# fsaverage white to .gii
convert_white = MRIsConvert()
convert_white.inputs.in_file = fsaverage_white
convert_white.inputs.out_file = "rh.white.surf.gii"
convert_white_results = convert_white.run()
fsaverage_white_gii = convert_white_results.outputs.get()['converted']
# Resample fsLr sphere to fsaverage
resample = LabelResampleTask()
resample.inputs.label_in = fs_LR_labels
resample.inputs.current_sphere = fs_LR_sphere
resample.inputs.new_sphere = fsaverage_sphere_gii
resample.inputs.method = "BARYCENTRIC"
resample.inputs.label_out = "sub-fsaverage_R.subparc374.label.gii"
resample_results = resample.run()
label_gii = resample_results.outputs.get()['out']
# fs average label.gii to .annot
convert_label = MRIsConvert()
convert_label.inputs.in_file = fsaverage_sphere_gii
convert_label.inputs.annot_file = label_gii
convert_label.inputs.out_file = 'sub-fsaverage_R.subparc374.annot'
convert_label_results = convert_label.run()
convert_fsaverage_annot = convert_label_results.outputs.get()['converted']
# mri_annotation2labe
surface_transform = SurfaceTransform()
surface_transform.inputs.hemi = 'rh'
surface_transform.inputs.target_subject = subject
surface_transform.inputs.source_subject = 'fsaverage'
surface_transform.inputs.source_annot_file = convert_fsaverage_annot
surface_transform.inputs.subjects_dir = subject_dir
surface_transform_results = surface_transform.run()
subject_annot = surface_transform_results.outputs.get()['out_file']
where LabelResampleTask is a simple wrapper around wb_command -label-resample command ( here ).
It seems that I’m missing a step to align fsLR sphere to fsaverage sphere, but can’t find a way to do it… How could I compute and apply such transformation ?
Or maybe it is a completely different issue ?
Version:
Environment (Docker, Singularity / Apptainer, custom installation):
Here is the neurodocker command to generate my dev environment:
neurodocker generate docker --base-image ubuntu:20.04 --env DEBIAN_FRONTEND=noninteractive --pkg-manager apt --run "export LD_LIBRARY_PATH=/lib64/:${LD_LIBRARY_PATH}" --install connectome-workbench --install graphviz xvfb qt5-default --freesurfer version=7.3.1 --miniconda version=latest conda_install="python=3.9 traits nipype==1.8.6 mne notebook vtk pyvista pyvistaqt pyqt matplotlib" --env MNE_3D_BACKEND=pyvista --env MNE_3D_OPTION_ANTIALIAS=false --env START_XVFB=true | docker build -t meg_fs_wb:7.3.1 -
Screen
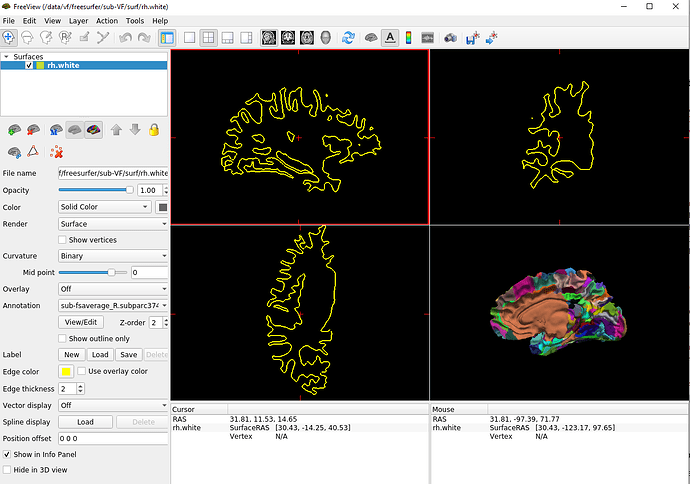
Here is the display of my freeview window with sub-VF rh.white + the output .annot file
We can clearly see that the medial wall is wrong.
Thanks for your time and help,
Would be happy to provide more details if needed