Long story short, I’m having issues with this registration. The resulting non-identity lta file will lead to a wrong aseg, which generates a wrong refined brain mask.
I went to the working directory and saw that this step uses mri_robust_register to register freesurfer/mri/T1.mgz to sub-xxx_T1w.nii.gz, showing one example below:
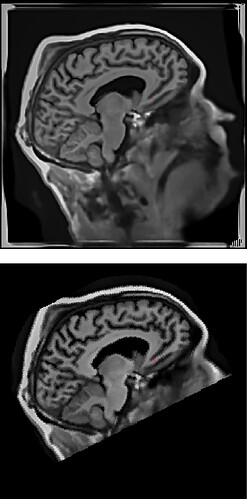
T1.mgz and sub-xxx_T1w.nii.gz look like the following:
I don’t think I can let mri_robust_register generate identity transformation (it should) by tuning its parameters given the truncation of the neck and those highlighted boundaries.
I came across this post. Just curious any reason not to assume the transform is identity as FreeSurfer always work in the native space, even when users pre-run it separately?
Back to this question itself, is there a flag that can be used to skip this registration step or simply force it to use identity?
Thanks,
Andrew
The motivating use case was that when you have multiple T1w images, a FreeSurfer user might combine them in one order and fMRIPrep might combine them in another. In the case of sub-01_ses-pre_T1w.nii.gz and sub-01_ses-post_T1w.nii.gz, the likely behavior was that the user would pass pre and then post, aligning to pre, but fMRIPrep, sorting lexically would align to post.
Thanks @effigies!! That makes perfect sense. Can I force it to identity (maybe through some flag) if I’m only using one T1w image and 100% sure they were aligned already?
Unfortunately not, at present. I wonder if we should register to rawavg.mgz instead, as it hasn’t had anything else done to it apart from converting to MGZ and averaging (copying for one image). Would you mind running mri_robust_register to rawavg.mgz and see if you get a reasonably small transform?
I have run several subjects in my database. rawavg.mgz works as expected with a standard freesurfer pipeline because, as you said, it does nothing except for file conversion and averaging (copying if there is only one t1w image).
However, just for your information, for many clinical cases where I need to run samseg and synthsr together with freesurfer, this still failed because the first step of samseg is conversion + conform. The conform turns the input image into a 256^3 image with 1mm isotropic resolution and cropping can happen near the borders where the highlights are as shown in the figures above. This is very specific to my cases. I will ping fs team for further discussion.
Nevertheless, I think replacing the source image from T1.mgz with rawavg.mgz in this registration step is in general a good move. It would be beneficial to provide such a flag in the future to force identity transform for users who know what they are doing (as you never know what images users could feed into the pipeline and what fs customization they have done  )
)
Thanks,
Andrew