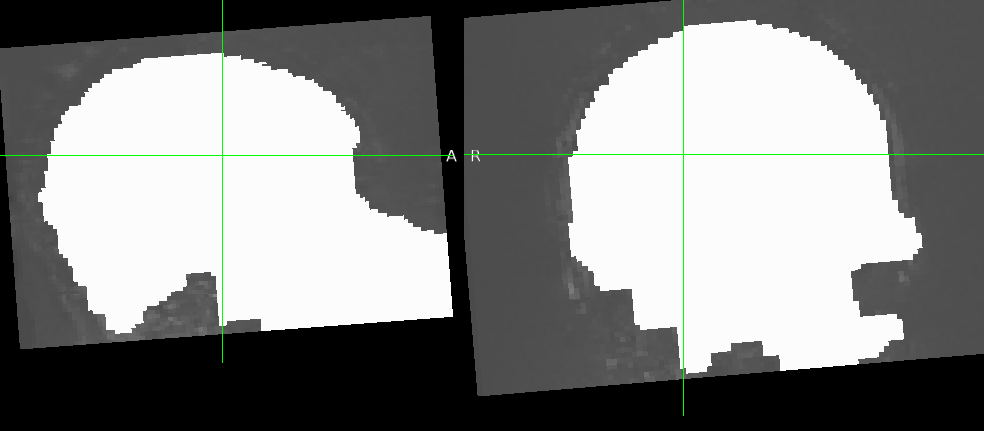
Summary of what happened:
Hello, we ran fmriprep on 49 subjects and for some of them the functional brain masks contain a significant amount of non-brain tissue, but the anatomical brain masks look fine. I am not sure why this happens, I have read it can be due to noisy background for example, but I don’t know if this is the case. Is there anything I can do in fmriprep, or outside and then feed it to fmriprep, to improve the bold brain mask generation?
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv \
-B ${DATA_DIR}:/data \
-B ${OUT_DIR}/:/out \
-B ${SCRATCH_DIR}:/wd \
-B ${LICENSE}:/license \
/fmriprep_singularity/fmriprep_25.1.3.simg \
--skip_bids_validation \
--participant-label ${SUBJECT} \
--omp-nthreads 5 --nthreads 5 --mem_mb 30000 \
--output-spaces MNI152NLin2009cAsym fsaverage\
--dummy-scans 5 \
--fs-license-file /license \
--work-dir /wd \
--cifti-output 91k \
/data /out/ participant
Version:
fMRIprep version 25.1.3
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS
Relevant log outputs (up to 20 lines):
NA
Screenshots / relevant information:
Example screenshot of the functional brain mask.