Hello, I’m using fmriprep on my data but my functional output is not aligned to MNI space, do you know how can I fix that?
Hi, to be able to help you, we would need more information:
- fmriprep version used
- fmriprep command used
- did the workflow execute cleanly or did you have any errors reported
- how did you see that the functional output is not aligned to MNI space…
In brief, all the points that are listed in the bug template from FMRIPREP bug report: https://github.com/nipreps/fmriprep/issues/new?assignees=&labels=bug&template=bug_report.yml
It is also worth noting that there are multiple MNI spaces. FSL’s default is the 6th generation Nonlinear Asym, while fMRIPrep’s default is 2009c nonlinear asym.
I’m using fmriprep 21.0.1
my command: singularity run -B /scratch --cleanenv fmriprep-21.0.1.simg /scratch/mahsa96/PT_PL029_V1 /scratch/mahsa96/PT_PL029_V1/derivatives participant --participant-label 01
–skip-bids-validation
–md-only-boilerplate
–fs-license-file /scratch/PT_PL029_V1/derivatives/license.txt
–fs-no-reconall
–output-spaces MNI152NLin2009cAsym:res-2
–stop-on-first-crash
–nthreads 32
–mem_mb 128000
I’m not sure what do you mean in your third question.
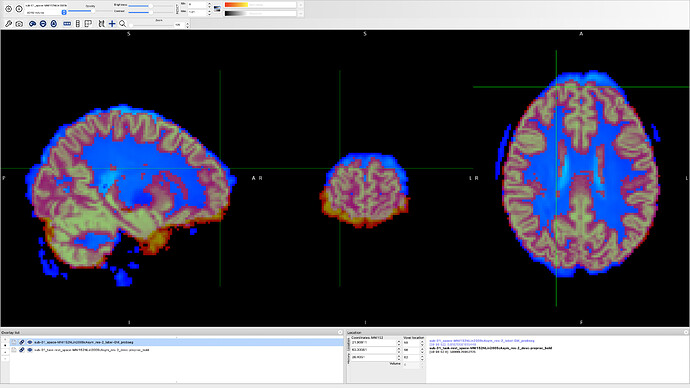
Actually, my GM output is not aligned to my bold output so I assumed that they are not aligned to MNI space.
Were there any errors in either the log file or the HTML summary file created by fMRIPrep?
Have you tried including freesurfer? Freesurfer boundary based registration tends to give a better performance than the alternative registration workflows.
Thank you for these precisions. Indeed the overlap between the functional image and the anatomical image does not look perfect. Do you have a fieldmap for this data set and was SDC performed on the functional image?
I see several options to try to change the result and to get a better overlap:
- –bold2t1w-dof : you can use 9 or 12 (default is 6). This may be relevant especially if your bold image was not corrected for susceptibility distorsions.
- –force-no-bbr or –force-bbr : this will change the heuristic used by fmriprep. I am not sure of this one.
- remove: -fs-no-reconall to use freesurfer and bbr for bold-to-T1w registration. This last option is also compatible with the variations above.
I tried actvating freesurfer and coregistration 12 but did not help! seriously I dont know what else I can do!
- When you re-ran fmriprep, did you start from a clean working directory? (according to your command line above I guess you did since you did not specify an explicit working directory with the -w option). You should consider specifying explicitly a location for your working directory, it will save the temporary files from fmriprep previous executions and in some cases FMRIPREP does not need to re-run everything again when you try different options.
- It would be interesting to look at the contrast in your T1w and BOLD images. Indeed the registration is based on the
bbr(boundary based registration) method (either from FSL or freesurfer), which implied that the software need to recognize the border between grey and white matter in both images. - Do you have SBref images saved by the scanner for reference of your bold image? It typically comes out from the bold sequences using simultaneous multi-slice acquisitions.
- Does fmriprep do SDC (susceptibility distorsion correction) for you? Do you provide a fmap for your bold runs where you use the ‘Intendedfor’ field in the json file to specify to which bold run SDC should be applied?
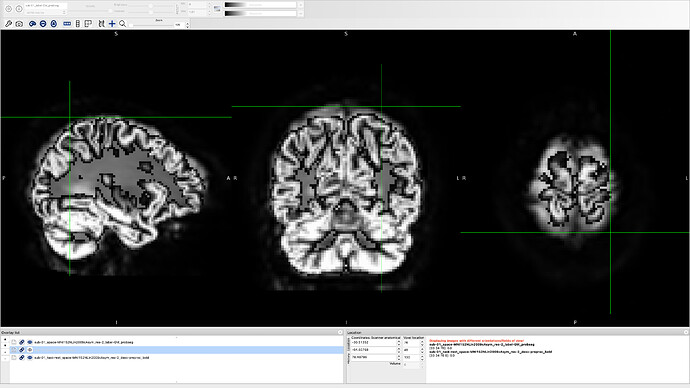
- It would be useful for us to get another view of your bold and T1w images, with a greyscale colormap for instance. Eventually, it would be useful to look at the flickering images of the FMRIPREP report to better assess how wrong is the registration/normalisation step.
yes, I re-ran it in a new work directory. and my data has fmap folder and every needed field in json file, about sbref, I don’t have that among my data.
I provide you with another image just to be clear.
Maybe what you see is in fact as good as the registration can get. Do not forget that on the bold image you see the CSF whose localization goes outside the cortical Gray Matter. When you look at the HTML report, does the registration look off to you?
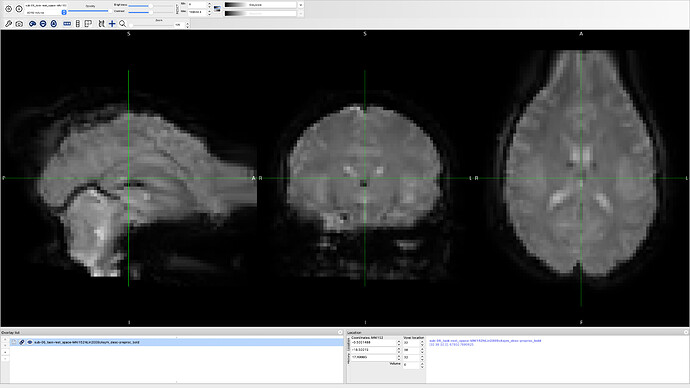
Well actually I removed output-spaces MNI152NLin2009cAsym:res-2 from my command line and now it is working just fine, but I have another problem, something weird! can you help me with that?
Is this still without using freesurfer? If so, can you try enabling freesurfer?
It looks like a susceptibility distortion correction step that went wrong. Is the phase encoding direction well described in the json file of the func/ files and the fmap/ files?
Based on what I understood, epi image and bold image should have opposite signs is it true? cause my scanner is Philips and I don’t know whether they are + or -. so I tested epi + , bold - and epi - , bold + in seprate runs but I have the same result!
I used freesurfe in my previous runs, I think this problem is related to my fmap and freesurfer is related to t1 analyze, is it true?
Yes Freesurfer is used on T1 images, but outputs from Freesurfer can inform spatial normalization. I agree this looks like a problem with susceptibility distortion correction, but just wanted to make sure it wasn’t suboptimal spatial normalization
I remove what I said. @Steven may be right with the non-linear transformation used in the normalization process being the culprit here. The shape of the artefact is much too strong and too localized (only the frontal part) to be due to the SDC process being wrong. I remember seeing this kind of dilation in my MRIQc report using partial coverage acquisition that were over dilated to fit a whole brain template. It may be the multiple degree of freedom of the non linear registration that went wrong. You could verify that by looking at the intermediate linearly registered bold image in the T1w space. If it is SDC that is wrong, you would see that in the T1w space. If you only see that in the Normalized space then it is the non linear registration that is wrong.