Summary of what happened:
Hey fmriprep-ers,
Would appreciate any help/guidance you can offer!
I have some multi-band fMRI data with a corresponding field map (in Hz) directly generated from GE scanner (as well as the accompanying magnitude file).
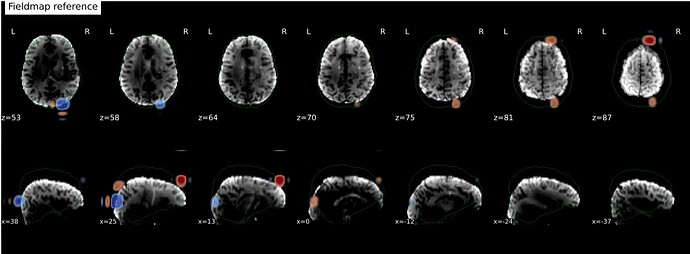
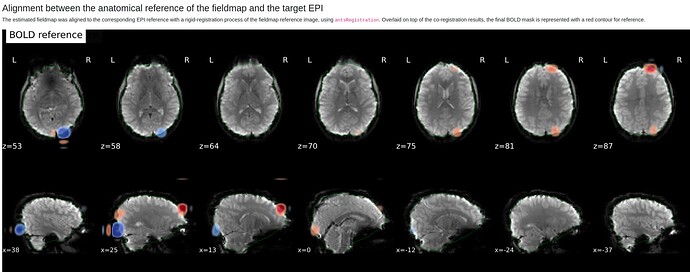
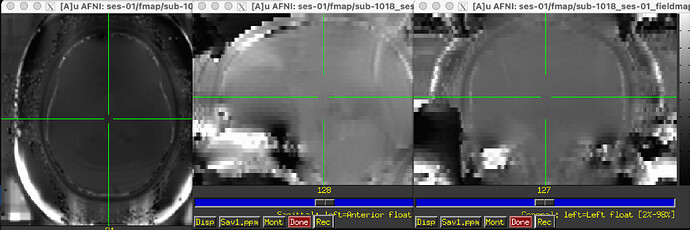
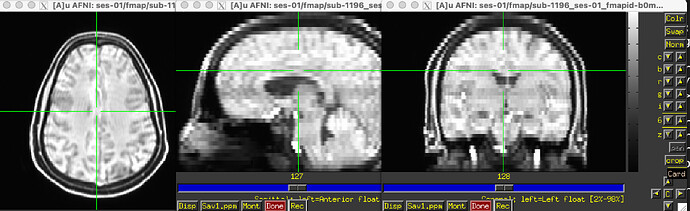
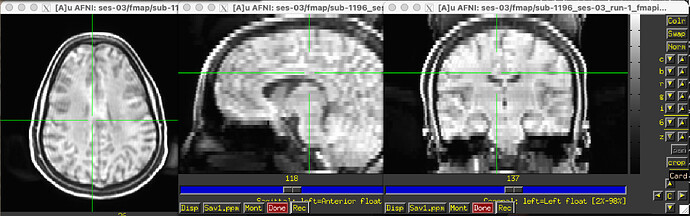
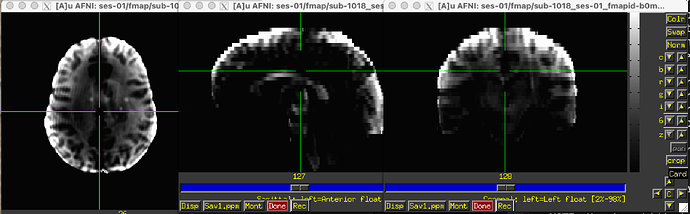
The odd part here is the major offset of the fieldmap and EPI target images (showing only the fieldmap reference; says I am only allowed to embed one media image… happy to add the BOLD reference image, examples of the raw fieldmap/magnitude files, and/or others to the comments, if I can)
For context, the original magnitude files had a ghosting issue (which was negatively impacting the SDC in FMRIPREP), our MR Physicist recommended prepending SSWarp to skull strip the ghosting out of the magnitude files prior to running FMRIPrep.
Perhaps this is what is causing the issue, open to other suggestions on how to handle the ghosting! The problem was fixed ~half way through data collection, so this is only impacting half our scans. Will try to add before/after of this step in comments, if possible.
Command used (and if a helper script was used, a link to the helper script or the command generated):
fmriprep-docker /data /out --fs-license-file /usr/local/freesurfer-current/license.txt --longitudinal --participant-label 1018
Version:
version = “23.2.0”
Environment (Docker, Singularity / Apptainer, custom installation):
Docker;
docker run --rm -e DOCKER_VERSION_8395080871=25.0.3 -it -v /usr/local/freesurfer-current/license.txt:/opt/freesurfer/license.txt:ro -v /Volumes/Vol6/LOKI/Imaging_Data/BIDS_Raw:/data:ro -v /Volumes/Vol6/LOKI/Imaging_Analyses/TD_Task_Analyses/:/out nipreps/fmriprep:23.2.0 /data /out participant --longitudinal --participant-label 1018
Data formatted according to a validatable standard? Please provide the output of the validator:
docker run -ti --rm -v /Volumes/Vol6/LOKI/Imaging_Data/BIDS_Raw/:/data:ro bids/validator /data
bids-validator@1.14.0
(node:1) Warning: Closing directory handle on garbage collection
1: [WARN] Not all subjects contain the same sessions. (code: 97 - MISSING_SESSION)
./sub-1185/ses-03
Evidence: Subject: sub-1185; Missing session: ses-03
./sub-1229/ses-02
Evidence: Subject: sub-1229; Missing session: ses-02
./sub-1229/ses-03
Evidence: Subject: sub-1229; Missing session: ses-03
./sub-1287/ses-02
Evidence: Subject: sub-1287; Missing session: ses-02
./sub-1287/ses-03
Evidence: Subject: sub-1287; Missing session: ses-03
Please visit https://neurostars.org/search?q=MISSING_SESSION for existing conversations about this issue.
2: [WARN] The Authors field of dataset_description.json should contain an array of fields - with one author per field. This was triggered based on the presence of only one author field. Please ignore if all contributors are already properly listed. (code: 102 - TOO_FEW_AUTHORS)
Please visit https://neurostars.org/search?q=TOO_FEW_AUTHORS for existing conversations about this issue.
Summary: Available Tasks: Available Modalities:
3738 Files, 133.52GB habi MRI
79 - Subjects cond
3 - Sessions vext
dext
rest
reca
rein
resting
If you have any questions, please post on https://neurostars.org/tags/bids.
Relevant log outputs (up to 20 lines):
No errors; the only log file included (fmriprep.toml). Here is part of the [workflow], happy to provide any other info.
anat_only = false
aroma_err_on_warn = false
aroma_melodic_dim = 0
bold2t1w_dof = 6
bold2t1w_init = "register"
cifti_output = false
fmap_bspline = false
force_syn = false
hires = true
ignore = []
level = "full"
longitudinal = true
run_msmsulc = true
medial_surface_nan = false
project_goodvoxels = false
regressors_all_comps = false
regressors_dvars_th = 1.5
regressors_fd_th = 0.5
run_reconall = true
skull_strip_fixed_seed = false
skull_strip_template = "OASIS30ANTs"
skull_strip_t1w = "force"
slice_time_ref = 0.5
spaces = "MNI152NLin2009cAsym:res-native"
use_aroma = false
use_syn_sdc = false
me_t2s_fit_method = "curvefit"
Screenshots / relevant information:
The folder structure is:
└── ses-01
├── anat
│ ├── sub-1260_ses-01_acq-MPRAGE_T1w.json
│ └── sub-1260_ses-01_acq-MPRAGE_T1w.nii.gz
├── fmap
│ ├── sub-1260_ses-01_fieldmap.json
│ ├── sub-1260_ses-01_fieldmap.nii.gz
│ └── sub-1260_ses-01_magnitude.json
│ ├── sub-1260_ses-01_magnitude.nii.gz
└── func
├── sub-1260_ses-01_task-cond_bold.json
├── sub-1260_ses-01_task-cond_bold.nii.gz
├── sub-1260_ses-01_task-cond_events.tsv
├── sub-1260_ses-01_task-habi_bold.json
└──sub-1260_ses-01_task-habi_bold.nii.gz
├──sub-1260_ses-01_task-habi_events.tsv
In the summary report, it mentions using:
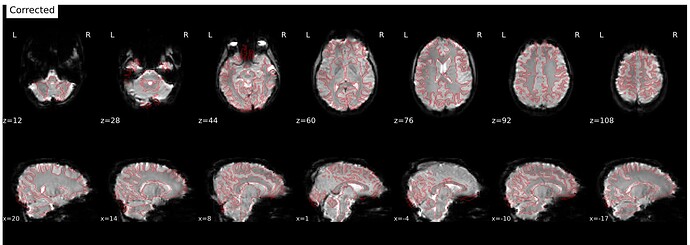
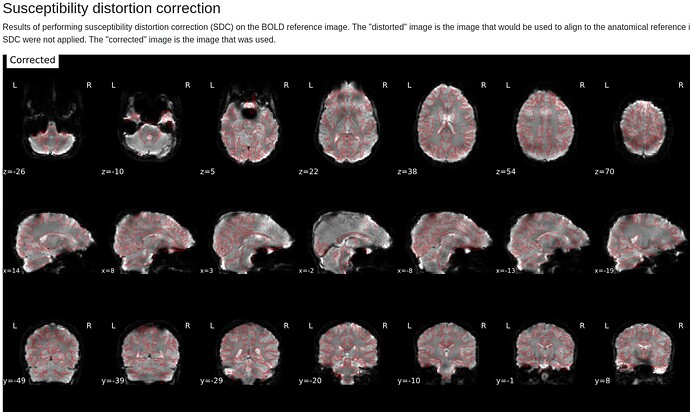
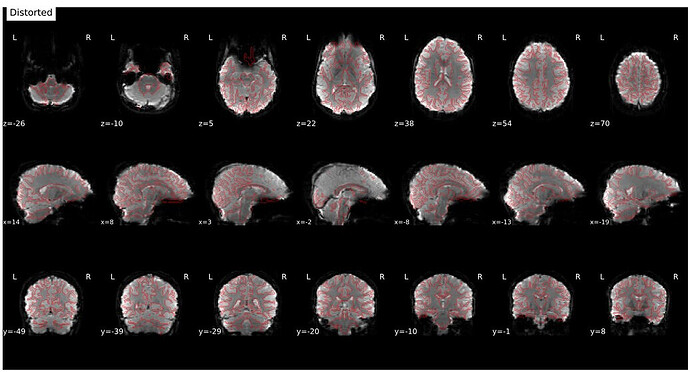
Susceptibility distortion correction: FMB (fieldmap-based) - directly measured B0 map
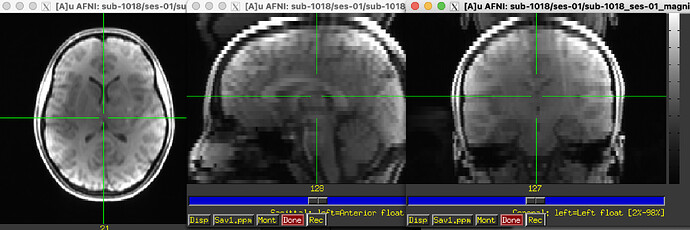
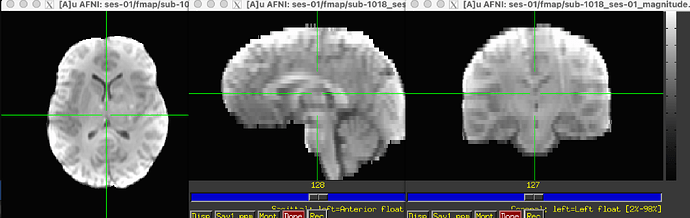
It’s a bit hard to see in the screenshot, but in the fmap reference, you can see the green outline is shifted up, as compared to the fmap. The SDC seemed to do pretty much nothing at all… the “distorted” and “corrected” images are essentially the same, with the only obvious difference bing the corrected images are brighter, per se. Will try to add images in a reply.