Summary of what happened:
I am validating some of the ROIs from my study but I got some weird results while computing the reverse inference when using the NiMARE - neurosynth_decode function.
Then I took core memory ROI i.e Hippocampus, with MNI coordinates (‘x’:-20,‘y’:-20,‘z’:-15) for test purpose but still getting the same results against the reverse inference (pReverse , zReverse, probReverse).
Can anyone point out the mistake - I am using just the MNI xyz coordinates for validating.
Python Code:
from nimare.extract import fetch_neurosynth
from nimare.io import convert_neurosynth_to_dataset
files = fetch_neurosynth(path='.', version='7', source='abstract', vocab='terms')
ns_dset = convert_neurosynth_to_dataset(
coordinates_file=files[0]["coordinates"],
metadata_file=files[0]["metadata"],
annotations_files=files[0]["features"]
)
import pandas as pd
from nimare.decode.discrete import neurosynth_decode
from nimare.dataset import Dataset
coord_df = pd.DataFrame([
{'id':'Hippocampus','x':-20,'y':-20,'z':-15},
{'id':'PFC','x':-45,'y':45,'z':15},
])
roi_ids = ['Hippocampus','PFC']
roi2studies = {}
for _, row in coord_df.iterrows():
matching_ids = ns_dset.get_studies_by_coordinate(
xyz=[(row.x, row.y, row.z)], # list of 3-tuples → shape (1, 3)
r=10.0 # radius in mm
)
roi2studies[row.id] = matching_ids
#---------------------------
study_ids = roi2studies['Hippocampus']
sub_dset = ns_dset.slice(study_ids) # keep only selected studies
# Broader memory terms that might exist
broader_memory_terms = [col for col in ns_dset.annotations.columns
if any(word in col.lower() for word in
['memory', 'recall', 'recognition', 'retrieval', 'encoding'])]
decoded_memory = neurosynth_decode(
coordinates=sub_dset.coordinates,
annotations=ns_dset.annotations,
ids=study_ids,
feature_group="terms_abstract_tfidf",
features=broader_memory_terms,
frequency_threshold=0.001,
correction="fdr_bh" # keep multiple-comparison control
)
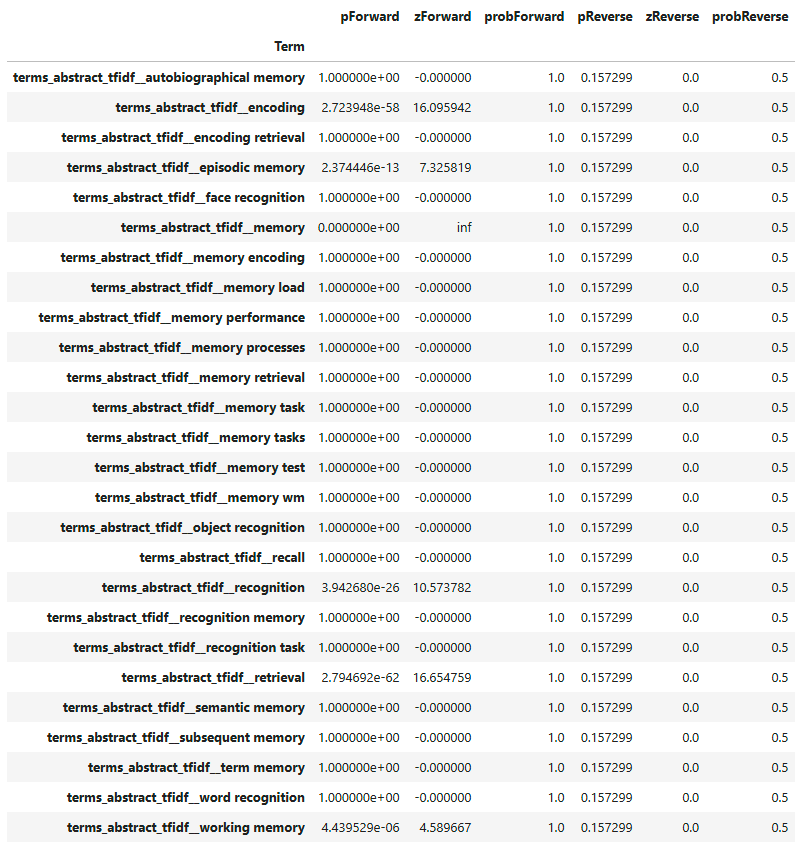
decoded_memory
Version:
My NiMARE version is ‘0.5.4’
Environment:
I have python 3.10.18 and using jupyter notebook and NiMARE version is ‘0.5.4’