I am aiming to combine some fsLR 32k surfaces I made from fmriprep outputs using this surface pipeline and these post-processing pipelines (anat space & MNI space) with subcortical data in MNI152NLin6Asym space (also postprocessed from fmriprep outputs).
This first step, I believe, is to grab just the subcortical data from the MNI outputs I have using the following command:
wb_command -cifti-create-dense-from-template \
91282_Greyordinates.dscalar.nii \
sub-MWMH317_ses-1_task-rest_space-fsLR_desc-subcort_bold.dscalar.nii \
-volume-all sub-MWMH317_ses-1_task-rest_space-MNI152NLin6Asym_desc-postproc_bold.nii.gz
When I run this command, however, I get the following error:
“ERROR: -volume-all specifies a volume file that doesn’t match the volume space of the template cifti file”
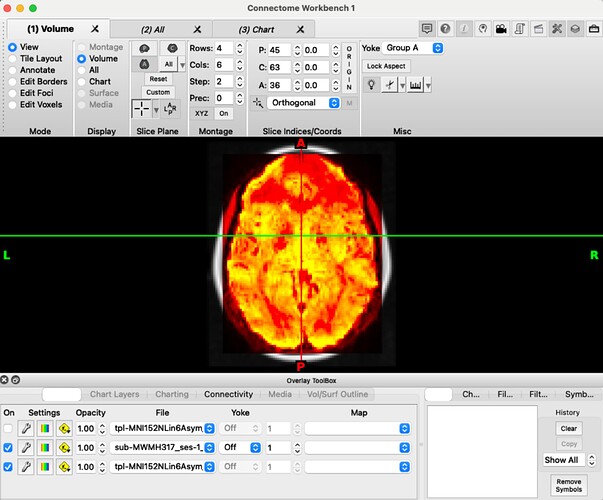
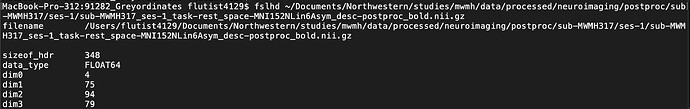
I looked into the nifti headers, and tried viewing the images, and it looks like while sub-MWMH317_ses-1_task-rest_space-MNI152NLin6Asym_desc-postproc_bold.nii.gz is in MNI152NLin6Asym space:
wb_view sub-MWMH317_ses-1_task-rest_space-MNI152NLin6Asym_desc-preproc_bold.nii.gz \
tpl-MNI152NLin6Asym_res-02_T1w.nii.gz
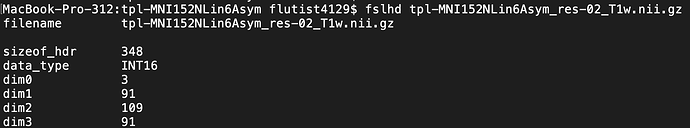
The images do not have the same dimensions:
I also double checked that tpl-MNI152NLin6Asym_res-02_T1w.nii.gz and 91282_Greyordinates.dscalar.nii are in the same space (via viewing) and have the same dimensions (via fslhd), and they do.
So I think the problem is that the fmriprep output has gotten rid of some voxels around the brain. Does anyone have suggestions for how I could address this issue? My first instinct is that I could add padding to match the fmriprep output to the MNI152NLin6Asym space, but I am not sure how to do this, and I am not sure if it is the best path forward.