Hello,
Main question: Does fmriprep handle gibbs ringing artifacts?
Issue at hand and background:
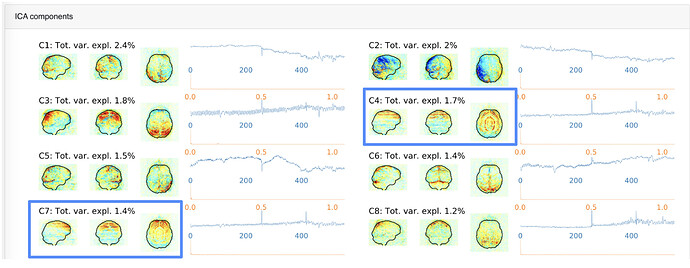
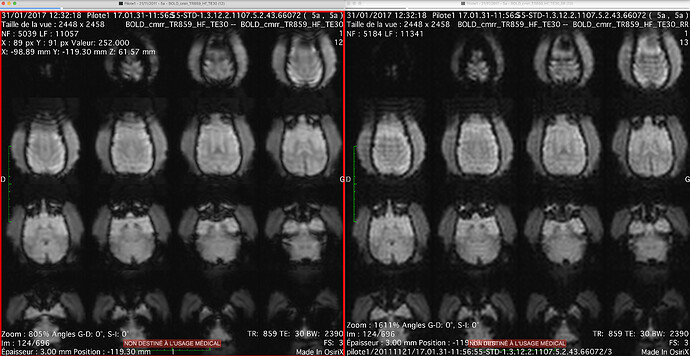
- We’ve noticed gibbs ringing within our mriqc outputs [ Figure 1 below ]
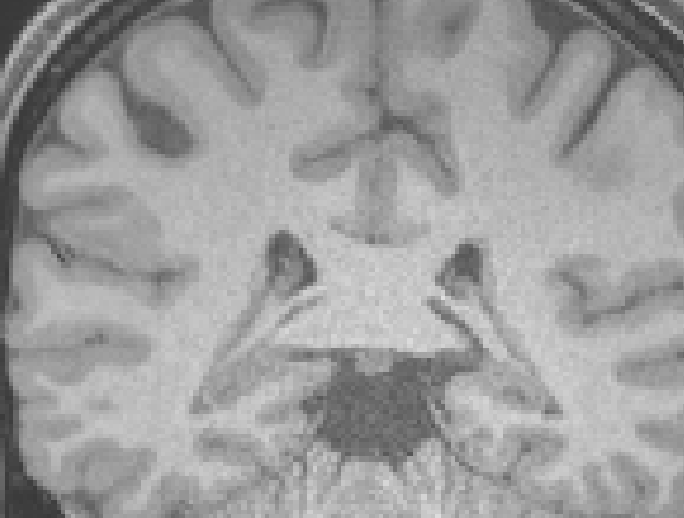
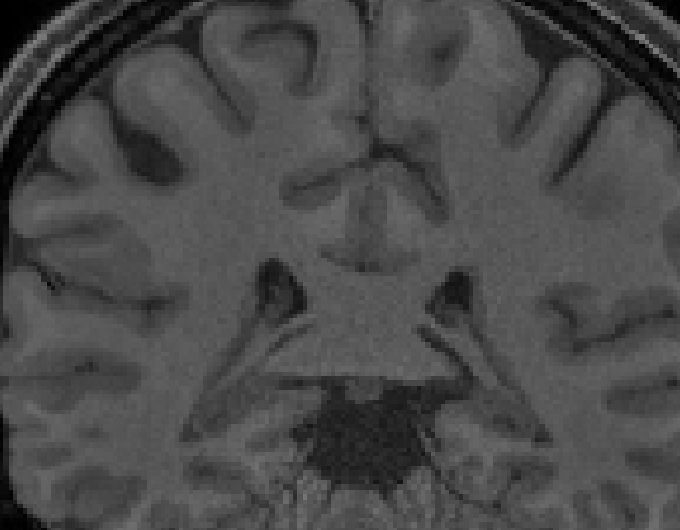
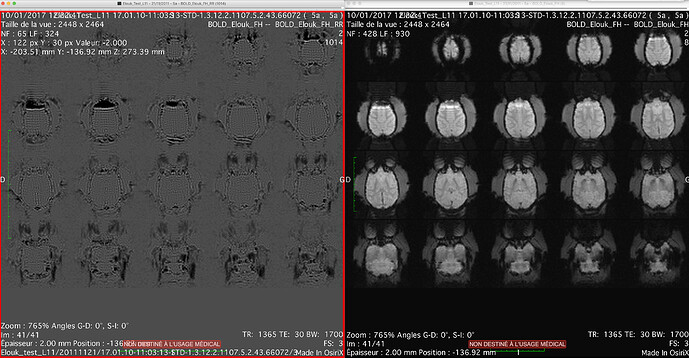
- The rings are still present in T1 images, after fmriprep preprocessing [ Figure 2 & 3 below ]
- Several communities have suggested that smoothing will resolve the gibbs artifact; however, we plan to NOT smooth the data for the purpose of running MVPA and hyperalignment.
- For DWI data, there are many resources regarding de-gibbs algorithms; we’re also aware that qsiprep has a degibbs option.
Back to question:
Does fmriprep handle gibbs artifacts? Or if we need to take additional steps, could you please recommend any “post” preprocessing algorithms, such as MRtrix3’s degibbs algorithm?
Thank you in advance!
Heejung
[ Figure 1: mriqc output, rings visible from the ICA components ]
[ Figure 2: T1w BEFORE fmriprep, rings visible ]
[ Figure 3: T1w AFTER fmriprep, rings visible ]