Mentors: Marcel Stimberg <marcel.stimberg@inserm.fr>, Dan Goodman <d.goodman@imperial.ac.uk>, Benjamin Evans <B.D.Evans@sussex.ac.uk>
Skill level: Intermediate/Advanced
Required skills: Python, C++
Time commitment: Full time (350 hours)
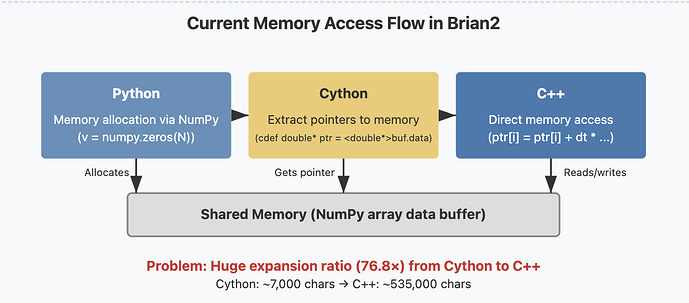
About: Brian is a clock-driven spiking neural network simulator that is easy to learn, highly flexible, and simple to extend. Written in Python, it allows users to describe and run arbitrary neural and synaptic models without needing to write code in any other programming language. It is built on a code-generation framework that transforms model descriptions into efficient low-level code. In Brian’s “runtime mode”, the generated code can interact with Python code by accessing the same data structures in memory. To make this work seamlessly, Brian makes use of Cython. This approach comes with two major disadvantages: 1) Cython compilation is slow (it generates a lot of code for error checking, etc.). For Cython, this is not a big downside, since it is commonly used to compile libraries once, but for Brian it matters since it needs to compile dynamically generated code frequently 2) We need to maintain a third code generation target besides Python and C++, with small but non-trivial differences to both of them.
Aims: The aims of this project are to:
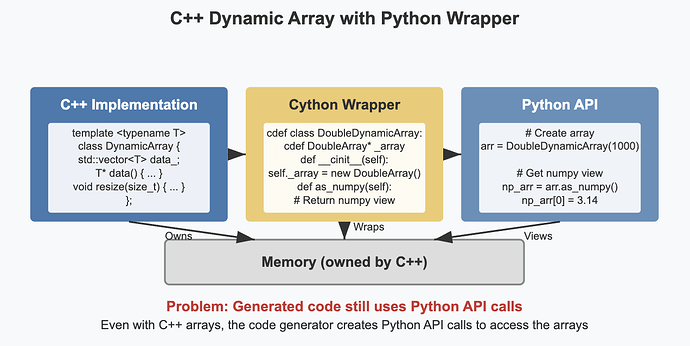
- Replace the Python data structures that are currently used from within Cython code (dynamic arrays and the “spike queue”) by C++ equivalents
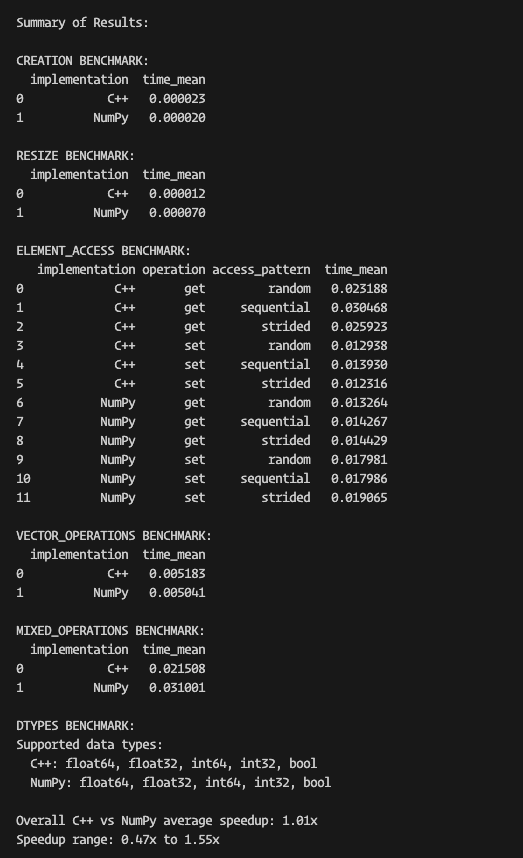
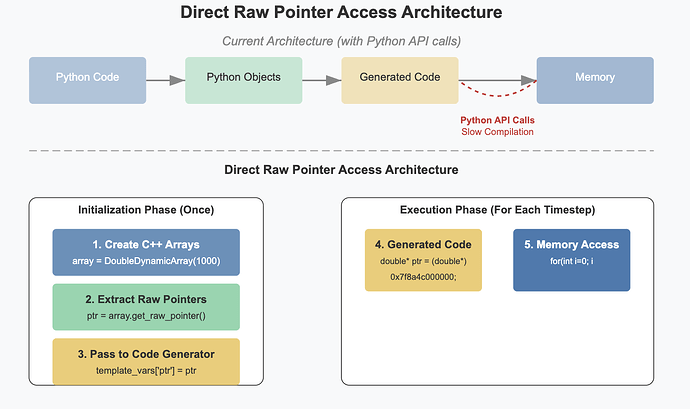
- Research solutions to call the compiled C++ code from Python and make it directly access the memory in the shared data structures storing the simulation variables. This could build upon existing just-in-time compilation technologies such as numba, or a package such as scipy-weave.
- Implement the above solution, and refactor the current code to make use of it
Project website: GitHub - brian-team/brian2: Brian is a free, open source simulator for spiking neural networks.
Tech keywords: Python, C++, compilation, JIT