Summary of what happened:
I am using fMRIPrep to preprocess resting-state data for an older-adult sample, some of whom have extensive age-related white matter lesions.
My understanding is that fMRIPrep uses FSL FAST to segment tissue types and create tissue probability maps, and these tissue masks are used to extract some of the confound timeseries (e.g. global_signal, csf, white_matter, a_comp_cor, etc).
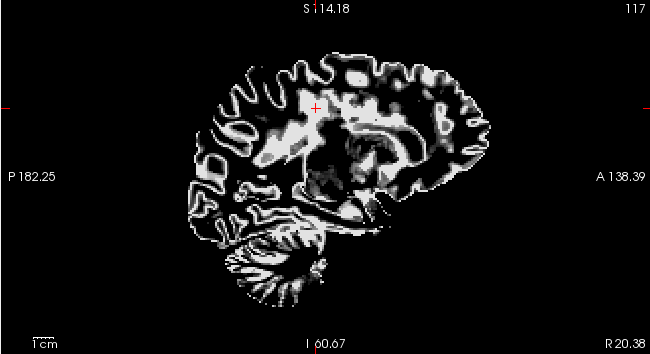
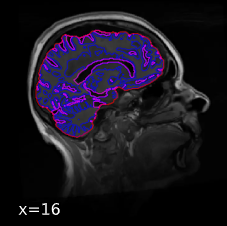
My problem is that FSL FAST misidentifies white matter lesions as gray matter, such that these lesions are included in the sub-*_label-GM_probseg.nii.gz files (see screenshots). The tissue-relevant confound timeseries are therefore being extracted from inaccurate tissue masks.
My questions:
-
Are there any existing functions/options in fMRIPrep that would enable me to address this issue? I would imagine any sizeable sample of older adults would run into this problem.
-
If answer to #1 is “no”, would the best course of action be to manually correct tissue masks by creating white matter lesion masks (using LST-AI or similar), subtracting them from the label-GM_probseg.nii.gz files (and maybe adding them into the label-WM_probseg.nii.gz files), and then re-create the confound timeseries on my own using the corrected masks?
-
Another option would be to use lesion filling/painting methods developed for imaging of multiple sclerosis patients, and supplying the lesion-filled T1s as input to fMRIPrep (although I know fMRIPrep requires/prefers to have the “raw” T1 as input), to increase the probability of these lesions being accurately segmented. A reasonable option, or are there downstream consequences I’ve missed?
-
Beyond the confound timeseries, are there additional consequences of the white matter lesion misclassification to consider? I’m trying to think through if it would have any impact on normalization, etc, and I don’t think it would.
Thanks in advance for any insight. I’m hoping someone with more experience/imagination than myself has already thought through this problem.
(Not sure if this belongs in Software Support or not, as it pertains to fMRIPrep software but is not a bug per se. Please let me know if it would be better placed in another category and I can create a new Topic as needed. Also, I did not include logs or validator output as they do not seem to be relevant to the question, but can post if needed.)
Command used (and if a helper script was used, a link to the helper script or the command generated):
#!/bin/bash
ml singularity
subj="$1"
singularity run -e --bind [data_path]:/data \
--bind [output_path]:/out \
--bind [license_path]:/license \
[fmriprep.sif path] \
/data /out participant \
--participant_label ${subj} \
--output-spaces fsnative fsaverage anat:res-native MNI152NLin6Asym:res-2 --fd-spike-threshold 0.5 --dvars-spike-threshold 3 \
--fs-license-file /license/license.txt -w [scratch_path]
Version: 24.1.0
Environment (Docker, Singularity / Apptainer, custom installation): Singularity via HPC
Screenshots / relevant information: