Summary of what happened:
I have an AP and PA that were acquired prior to each func run, but fmriprep claims that only 1 fieldmap can be generated: A total of 1 fieldmaps were found available within the input BIDS structure for this particular subject. A B0-nonuniformity map (or fieldmap) was estimated based on two (or more) echo-planar imaging (EPI) references with topup (Andersson, Skare, and Ashburner (2003); FSL 6.0.5.1:57b01774). This is copied from the html.
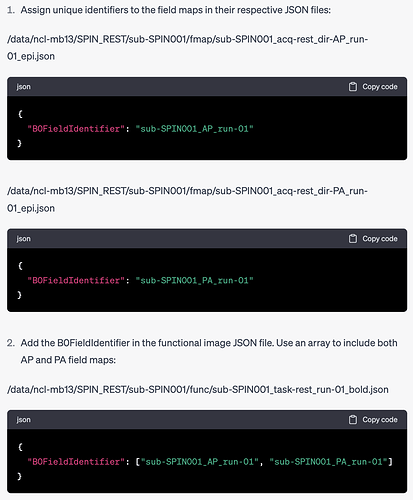
I have specified in each .json file of the AP and PA images which func run they are intended for like so: "IntendedFor": ["func/sub-SPIN001_task-rest_run-02_bold.nii.gz"],
Everything else with fmriprep seems quite perfect. I made sure to pass the bids-validator with no errors prior to running fmriprep.
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv -B /data $sif_loc $input_dir $output_dir participant --participant-label $subject --nthreads 16 --omp-nthreads 16 --mem_mb 32000 --skip-bids-validation --output-spaces MNIPediatricAsym:cohort-4:res-2 --use-aroma --skull-strip-template NKI --fs-license-file $fs_license --work-dir $work_dir
Version:
fMRIPrep v23.0.0rc0
Environment (Docker, Singularity, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
Yes. There is one strange warning:
1: [WARN] Not all subjects contain the same files. Each subject should contain the same number of files with the same naming unless some files are known to be missing. (code: 38 - INCONSISTENT_SUBJECTS)
./sub-SPIN001/fmap/sub-SPIN001_acq-rest_dir-AP_epi.json
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_acq-rest_dir-AP_epi.json
./sub-SPIN001/fmap/sub-SPIN001_acq-rest_dir-AP_epi.nii.gz
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_acq-rest_dir-AP_epi.nii.gz
./sub-SPIN001/fmap/sub-SPIN001_acq-rest_dir-AP_run-03_epi.json
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_acq-rest_dir-AP_run-03_epi.json
./sub-SPIN001/fmap/sub-SPIN001_acq-rest_dir-AP_run-03_epi.nii.gz
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_acq-rest_dir-AP_run-03_epi.nii.gz
./sub-SPIN001/fmap/sub-SPIN001_acq-rest_dir-PA_epi.json
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_acq-rest_dir-PA_epi.json
./sub-SPIN001/fmap/sub-SPIN001_acq-rest_dir-PA_epi.nii.gz
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_acq-rest_dir-PA_epi.nii.gz
./sub-SPIN001/fmap/sub-SPIN001_acq-rest_dir-PA_run-03_epi.json
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_acq-rest_dir-PA_run-03_epi.json
./sub-SPIN001/fmap/sub-SPIN001_acq-rest_dir-PA_run-03_epi.nii.gz
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_acq-rest_dir-PA_run-03_epi.nii.gz
./sub-SPIN001/func/sub-SPIN001_task-rest_bold.json
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_task-rest_bold.json
./sub-SPIN001/func/sub-SPIN001_task-rest_bold.nii.gz
Evidence: Subject: sub-SPIN001; Missing file: sub-SPIN001_task-rest_bold.nii.gz
... and 1592 more files having this issue (Use --verbose to see them all).
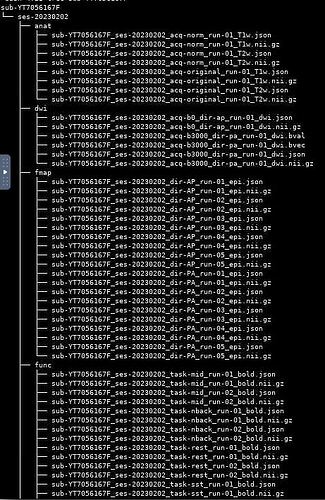
The above output is extremely weird because I don’t even have some of the above files. I did do some file cleaning/removing and ran the bids-validator after. The files should be:
ender.540> cd sub-SPIN001
ender.541> ls
anat fmap func
ender.542> ls -R
.:
anat fmap func
./anat:
sub-SPIN001_T1w.json sub-SPIN001_T1w.nii.gz
./fmap:
sub-SPIN001_acq-rest_dir-AP_run-01_epi.json sub-SPIN001_acq-rest_dir-PA_run-01_epi.json
sub-SPIN001_acq-rest_dir-AP_run-01_epi.nii.gz sub-SPIN001_acq-rest_dir-PA_run-01_epi.nii.gz
sub-SPIN001_acq-rest_dir-AP_run-02_epi.json sub-SPIN001_acq-rest_dir-PA_run-02_epi.json
sub-SPIN001_acq-rest_dir-AP_run-02_epi.nii.gz sub-SPIN001_acq-rest_dir-PA_run-02_epi.nii.gz
./func:
sub-SPIN001_task-rest_run-01_bold.json sub-SPIN001_task-rest_run-02_bold.json
sub-SPIN001_task-rest_run-01_bold.nii.gz sub-SPIN001_task-rest_run-02_bold.nii.gz
Relevant log outputs (up to 20 lines):
None.
Screenshots / relevant information:
None.