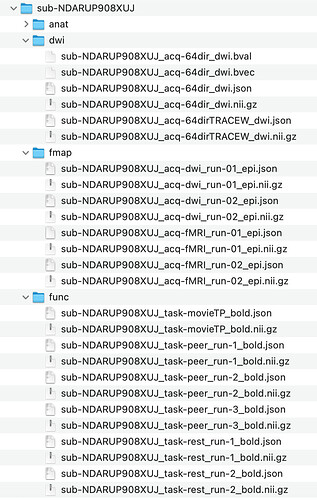
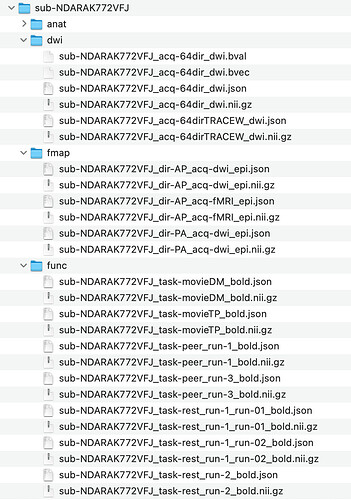
a bit more detailed examples :
ANAT:
.json: equal
’sub-NDARAU530GLJ_ses-SiteCBIC_acq-VNavNorm_run-01_T1w.json vs sub-NDARAU530GLJ_ses-SiteCBIC_acq-VNavNorm_run-02_T1w.json: equal’,
.json: ImageOrientationPatientDICOM
‘sub-NDARDN089RX5_ses-SiteCBIC_acq-HCP_run-02_T1w.json vs sub-NDARDN089RX5_ses-SiteCBIC_acq-HCP_run-01_T1w.json: ImageOrientationPatientDICOM: 3.04068e-05, 1, 2.64951e-08, 0.0487735, -1.45658e-06, -0.99881 != 0.0436194, 0.999048, 5.6545e-08, -0.0313809, 0.00137018, -0.999507; TxRefAmp: 211.304 != 212.794; SAR: 0.0903668 != 0.0927018’,
.json: TxRefAmp + SAR
sub-NDARUC591FWN_ses-SiteCBIC_acq-VNav_run-01_T1w.json vs sub-NDARUC591FWN_ses-SiteCBIC_acq-VNav_run-02_T1w.json: TxRefAmp: 212.955 != 213.947; SAR: 0.051552 != 0.0506172;
.json: TxRefAmp+ ReceiveCoilActiveElements + ReceiveCoilName + ShimSetting
‘sub-NDARVM557ZLF_ses-SiteCUNY_acq-VNavNorm_run-02_T1w.json vs sub-NDARVM557ZLF_ses-SiteCUNY_acq-VNavNorm_run-04_T1w.json: TxRefAmp: 242.478 != 237.848; ReceiveCoilActiveElements: BC != HEA;HEP; SAR: 0.144497 != 0.148159; ShimSetting: -1647, -10158, 2069, -170, 246, -36, 135, 37 != -1640, -10182, 2233, 354, 158, -117, 230, 25; ReceiveCoilName: Body != Head_32; InstitutionName missing in json1; InstitutionalDepartmentName missing in json1; InstitutionAddress missing in json1; SeriesNumber missing in json1; AcquisitionTime missing in json1; AcquisitionNumber missing in json1’,
.json: SAR
‘sub-NDARXB023AMW_ses-SiteRU_acq-HCP_run-02_T2w.json vs sub-NDARXB023AMW_ses-SiteRU_acq-HCP_run-01_T2w.json: SAR: 0.264022 != 0.267865’
FUNC:
.json: SliceTiming
‘sub-NDAREY381DEQ_ses-SiteCUNY_task-movieDM_run-02_bold.json vs sub-NDAREY381DEQ_ses-SiteCUNY_task-movieDM_run-01_bold.json: SliceTiming: 0, 0.545, 0.3125, 0.0775, 0.625, 0.39, 0.155, 0.7025, 0.4675, 0.235, 0, 0.545, 0.3125, 0.0775, 0.625, 0.39, 0.155, 0.7025, 0.4675, 0.235, 0, 0.545, 0.3125, 0.0775, 0.625, 0.39, 0.155, 0.7025, 0.4675, 0.235, 0, 0.545, 0.3125, 0.0775, 0.625, 0.39, 0.155, 0.7025, 0.4675, 0.235, 0, 0.545, 0.3125, 0.0775, 0.625, 0.39, 0.155, 0.7025, 0.4675, 0.235, 0, 0.545, 0.3125, 0.0775, 0.625, 0.39, 0.155, 0.7025, 0.4675, 0.235 != 0, 0.5475, 0.3125, 0.0775, 0.625, 0.39, 0.1575, 0.7025, 0.4675, 0.235, 0, 0.5475, 0.3125, 0.0775, 0.625, 0.39, 0.1575, 0.7025, 0.4675, 0.235, 0, 0.5475, 0.3125, 0.0775, 0.625, 0.39, 0.1575, 0.7025, 0.4675, 0.235, 0, 0.5475, 0.3125, 0.0775, 0.625, 0.39, 0.1575, 0.7025, 0.4675, 0.235, 0, 0.5475, 0.3125, 0.0775, 0.625, 0.39, 0.1575, 0.7025, 0.4675, 0.235, 0, 0.5475, 0.3125, 0.0775, 0.625, 0.39, 0.1575, 0.7025, 0.4675, 0.235’
DWI:
.json: equal
sub-NDARBF911MLX_ses-SiteCUNY_acq-dwi_dir-PA_run-01_epi.json vs sub-NDARBF911MLX_ses-SiteCUNY_acq-dwi_dir-PA_run-02_epi.json: equal’,
.json: SliceTiming+ImageOrientationPatientDICOM+TxRefAmp,
‘sub-NDARMH127CEW_ses-SiteCUNY_acq-64dir_run-01_dwi.json vs sub-NDARMH127CEW_ses-SiteCUNY_acq-64dir_run-02_dwi.json: ImageOrientationPatientDICOM: 0.993171, 0, 0.116671, 0, 1, 0 != 0.985991, 0.165048, 0.0240971, -0.164998, 0.986286, -0.00403247; TxRefAmp: 228.663 != 229.261; SAR: 0.525696 != 0.532975; ShimSetting: -1560, -10252, 2240, 757, 236, -277, 232, -77 != -1625, -10297, 2288, 719, 126, -401, 265, -64’
.json: SliceTiming,
'sub-NDARRN996JZK_ses-SiteCBIC_acq-64dir_run-01_dwi.json vs sub-NDARRN996JZK_ses-SiteCBIC_acq-64dir_run-02_dwi.json: SliceTiming: 0, 1.5125, 3.0225, 1.2375, 2.7475, 0.9625, 2.4725, 0.6875, 2.1975, 0.4125, 1.925, 0.1375, 1.65, 3.16, 1.375, 2.885, 1.1, 2.61, 0.825, 2.335, 0.55, 2.06, 0.275, 1.7875, 0, 1.5125, 3.0225, 1.2375, 2.7475, 0.9625, 2.4725, 0.6875, 2.1975, 0.4125, 1.925, 0.1375, 1.65, 3.16, 1.375, 2.885, 1.1, 2.61, 0.825, 2.335, 0.55, 2.06, 0.275, 1.7875, 0, 1.5125, 3.0225, 1.2375, 2.7475, 0.9625, 2.4725, 0.6875, 2.1975, 0.4125, 1.925, 0.1375, 1.65, 3.16, 1.375, 2.885, 1.1, 2.61, 0.825, 2.335, 0.55, 2.06, 0.275, 1.7875 != 0, 1.5125, 3.025, 1.2375, 2.75, 0.9625, 2.475, 0.6875, 2.2, 0.4125, 1.925, 0.1375, 1.65, 3.1625, 1.375, 2.8875, 1.1, 2.6125, 0.825, 2.3375, 0.55, 2.0625, 0.275, 1.7875, 0, 1.5125, 3.025, 1.2375, 2.75, 0.9625, 2.475, 0.6875, 2.2, 0.4125, 1.925, 0.1375, 1.65, 3.1625, 1.375, 2.8875, 1.1, 2.6125, 0.825, 2.3375, 0.55, 2.0625, 0.275, 1.7875, 0, 1.5125, 3.025, 1.2375, 2.75, 0.9625, 2.475, 0.6875, 2.2, 0.4125, 1.925, 0.1375, 1.65, 3.1625, 1.375, 2.8875, 1.1, 2.6125, 0.825, 2.3
FMAP:
.json: ShimSetting + SAR
'sub-NDARPH340DKQ_ses-SiteCUNY_acq-fMRI_dir-AP_run-02_epi.json vs sub-NDARPH340DKQ_ses-SiteCUNY_acq-fMRI_dir-AP_run-01_epi.json: SAR: 0.343403 != 0.339071; ShimSetting: -1592, -10184, 2273, 542, 14, -18, 242, -72 != -1559, -10161, 2246, 471, 102, -67, 222, -52; SliceTiming: 2.64, 0, 2.7275, 0.0875, 2.815, 0.175, 2.905, 0.2625, 2.9925, 0.3525, 3.08, 0.44, 3.1675, 0.5275, 3.255, 0.615, 3.345, 0.7025, 3.4325, 0.7925, 3.52, 0.88, 3.6075, 0.9675, 3.695, 1.055, 3.785, 1.1425, 3.8725, 1.2325, 3.96, 1.32, 4.0475, 1.4075, 4.135, 1.495, 4.225, 1.5825, 4.3125, 1.6725, 4.4, 1.76, 4.4875, 1.8475, 4.575, 1.935, 4.665, 2.0225, 4.7525, 2.1125, 4.84, 2.2, 4.9275, 2.2875, 5.015, 2.375, 5.105, 2.465, 5.1925, 2.5525 != 2.64, 0, 2.7275, 0.0875, 2.8175, 0.1775, 2.905, 0.265, 2.9925, 0.3525, 3.08, 0.44, 3.1675, 0.5275, 3.2575, 0.6175, 3.345, 0.705, 3.4325, 0.7925, 3.52, 0.88, 3.6075, 0.9675, 3.6975, 1.0575, 3.785, 1.145, 3.8725, 1.2325, 3.96, 1.32, 4.0475, 1.4075, 4.1375, 1.4975, 4.225, 1.585, 4.3125, 1.6725, 4.4, 1.76, 4.49, 1.8475, 4.5775, 1.9375, 4.665, 2.025, 4.7525, 2.1125, 4.84, 2.2, 4.93, 2.2875, 5.0175, 2.3775, 5.105, 2.465, 5.1925, 2.5525; TxRefAmp: 232.262 != 232.60
.json: ShimSetting
sub-NDARPJ502KEH_ses-Sit eCBIC_acq-fMRI_dir-PA_run-02_epi.json vs sub-NDARPJ502KEH_ses-SiteCBIC_acq-fMRI_dir-PA_run-01_epi.json: ShimSetting: 198, -10448, -5575, 494, -4, -163, 65, -12 != 200, -10475, -5596, 341, -8, -285, 96, -19; SliceTiming: 2.64, 0, 2.7275, 0.0875, 2.8175, 0.175, 2.905, 0.265, 2.9925, 0.3525, 3.08, 0.44, 3.1675, 0.5275, 3.2575, 0.615, 3.345, 0.705, 3.4325, 0.7925, 3.52, 0.88, 3.6075, 0.9675, 3.6975, 1.055, 3.785, 1.145, 3.8725, 1.2325, 3.96, 1.32, 4.0475, 1.4075, 4.1375, 1.495, 4.225, 1.585, 4.3125, 1.6725, 4.4, 1.76, 4.4875, 1.8475, 4.5775, 1.9375, 4.665, 2.025, 4.7525, 2.1125, 4.84, 2.2, 4.9275, 2.2875, 5.0175, 2.3775, 5.105, 2.465, 5.1925, 2.5525 != 2.64, 0, 2.7275, 0.0875, 2.815, 0.175, 2.9025, 0.2625, 2.9925, 0.3525, 3.08, 0.44, 3.1675, 0.5275, 3.255, 0.615, 3.3425, 0.7025, 3.4325, 0.7925, 3.52, 0.88, 3.6075, 0.9675, 3.695, 1.055, 3.7825, 1.1425, 3.8725, 1.2325, 3.96, 1.32, 4.0475, 1.4075, 4.135, 1.495, 4.2225, 1.5825, 4.3125, 1.6725, 4.4, 1.76, 4.4875, 1.8475, 4.575, 1.935, 4.665, 2.0225, 4.7525, 2.1125, 4.84, 2.2, 4.9275, 2.2875, 5.015, 2.375, 5.105, 2.4625, 5.1925, 2.55
25’,