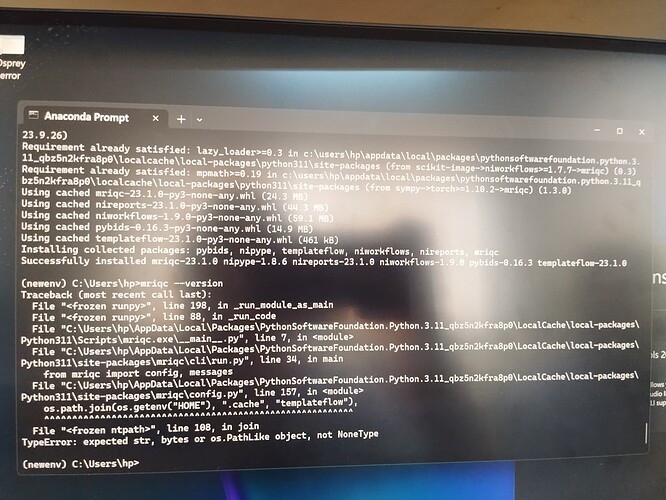
Hi @Steven,
Please find the output of the docker version below:
C:\Users\hp>docker run -it nipreps/mriqc:latest --version
MRIQC v23.1.0
C:\Users\hp>docker run -it --rm -v D:/test_29_final2:/data:ro -v D:/test_29_output:/out nipreps/mriqc:latest /data /out participant --participant_label 29
231120-06:39:35,810 cli IMPORTANT:
Running MRIQC version 23.1.0:
* BIDS dataset path: /data.
* Output folder: /out.
* Analysis levels: ['participant'].
231120-06:39:47,545 cli WARNING:
IMPORTANT: Anonymized quality metrics (IQMs) will be submitted to MRIQC's metrics repository. Submission of IQMs can be disabled using the ``--no-sub`` argument. Please visit https://mriqc.readthedocs.io/en/latest/dsa.html to revise MRIQC's Data Sharing Agreement.
231120-06:40:22,287 nipype.workflow WARNING:
Storing result file without outputs
231120-06:40:22,287 nipype.workflow WARNING:
[Node] Error on "mriqc_wf.dwiMRIQC.dwi_reference_wf.epi_merge" (/tmp/work/mriqc_wf/dwiMRIQC/dwi_reference_wf/_in_file_..data..sub-29..ses-001..dwi..sub-29_ses-001_acq-DTI_run-01_dwi.nii.gz/epi_merge)
231120-06:40:24,122 nipype.workflow ERROR:
Node epi_merge.a0 failed to run on host ff7758b6a615.
231120-06:40:24,128 nipype.workflow ERROR:
Saving crash info to /out/logs/crash-20231120-064024-root-epi_merge.a0-85ca863d-80f2-45a0-98b6-5e0109670366.txt
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/mriqc/engine/plugin.py", line 60, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node epi_merge.
Cmdline:
echo Only one time point!
Stdout:
Stderr:
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 752, in _run_interface
raise IOError(
OSError: No command "mri_robust_template" found on host ff7758b6a615. Please check that the corresponding package is installed.
Traceback (most recent call last):
File "/opt/conda/bin/mriqc", line 8, in <module>
sys.exit(main())
File "/opt/conda/lib/python3.9/site-packages/mriqc/cli/run.py", line 168, in main
mriqc_wf.run(**_plugin)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/workflows.py", line 638, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File "/opt/conda/lib/python3.9/site-packages/mriqc/engine/plugin.py", line 184, in run
self._clean_queue(jobid, graph, result=result)
File "/opt/conda/lib/python3.9/site-packages/mriqc/engine/plugin.py", line 256, in _clean_queue
raise RuntimeError("".join(result["traceback"]))
RuntimeError: Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/mriqc/engine/plugin.py", line 60, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node epi_merge.
Cmdline:
echo Only one time point!
Stdout:
Stderr:
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/opt/conda/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 752, in _run_interface
raise IOError(
OSError: No command "mri_robust_template" found on host ff7758b6a615. Please check that the corresponding package is installed.
Thanks & Regards,
Himanshu