Hi,
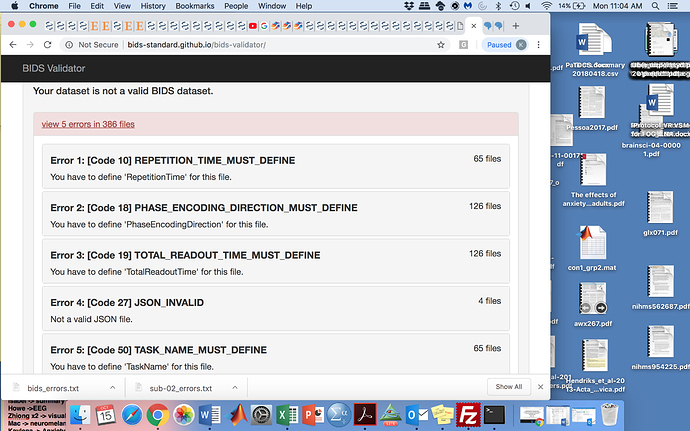
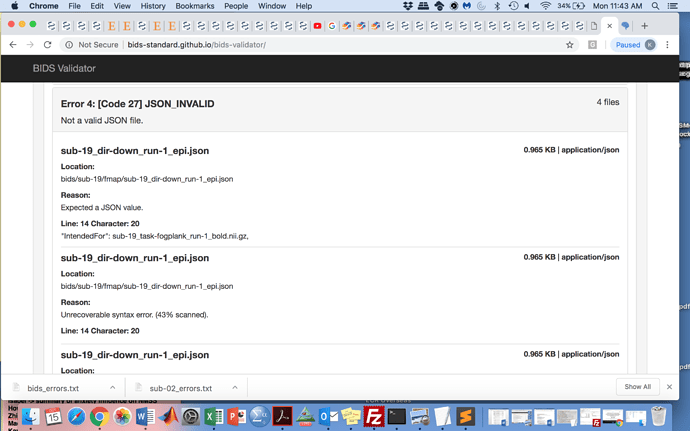
I am having trouble creating a bids valid dataset. I have used the dcm2nii to create the .json files and have adjusted the files to the bids specifications to the best of my knowledge, however many of the errors that are being flagged with the bids validator seem false. For example, repetition time, task name, etc are included in the json files.
I have tried to run fmriprep on a single subject, since I cannot find a way to make the dataset bids appropriate, however I get the following error:
Process Process-2:
Traceback (most recent call last):
File “/usr/local/miniconda/lib/python3.6/multiprocessing/process.py”, line 258, in _bootstrap
self.run()
File “/usr/local/miniconda/lib/python3.6/multiprocessing/process.py”, line 93, in run
self._target(*self._args, **self._kwargs)
File “/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/cli/run.py”, line 510, in build_workflow
ignore_aroma_err=opts.ignore_aroma_denoising_errors,
File “/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/workflows/base.py”, line 202, in init_fmriprep_wf
ignore_aroma_err=ignore_aroma_err)
File “/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/workflows/base.py”, line 458, in init_single_subject_wf
ignore_aroma_err=ignore_aroma_err)
File “/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/workflows/bold/base.py”, line 262, in init_func_preproc_wf
fmaps = layout.get_fieldmap(ref_file, return_list=True)
File “/usr/local/miniconda/lib/python3.6/site-packages/bids/grabbids/bids_layout.py”, line 244, in get_fieldmap
fieldmaps = self._get_fieldmaps(path)
File “/usr/local/miniconda/lib/python3.6/site-packages/bids/grabbids/bids_layout.py”, line 267, in _get_fieldmaps
metadata = self.get_metadata(file.filename)
File “/usr/local/miniconda/lib/python3.6/site-packages/bids/grabbids/bids_layout.py”, line 169, in get_metadata
encoding=‘utf-8’))
File “/usr/local/miniconda/lib/python3.6/json/init.py”, line 299, in load
parse_constant=parse_constant, object_pairs_hook=object_pairs_hook, **kw)
File “/usr/local/miniconda/lib/python3.6/json/init.py”, line 354, in loads
return _default_decoder.decode(s)
File “/usr/local/miniconda/lib/python3.6/json/decoder.py”, line 339, in decode
obj, end = self.raw_decode(s, idx=_w(s, 0).end())
File “/usr/local/miniconda/lib/python3.6/json/decoder.py”, line 357, in raw_decode
raise JSONDecodeError(“Expecting value”, s, err.value) from None
json.decoder.JSONDecodeError: Expecting value: line 14 column 17 (char 396)
Can anyone tell me what I am doing wrong?
Thanks,
Kaylena