Greetings,
I’m trying to learn about the freesurfer files produced by my fmriprep v20.2.7 pipeline.
My goal is to use the perirhinal label file generated by freesurfer and turn it into a native space binary mask.
My searches lead me to use mri_label2vol - Free Surfer Wiki function to transform my perirhinal_exvivo.thresh.label files to volumes.
import subprocess
# Define the subject's directory
SUBJECTS_DIR="/xxx/xxx/xxx/xxx/bids/derivatives/fmriprep-20.2.7"
# Define the subject ID
SUBJECT_ID="sub-xxx"
# Define the label paths
label_path_left = f"{SUBJECTS_DIR}/freesurfer/{SUBJECT_ID}/label/lh.perirhinal_exvivo.thresh.label"
label_path_right = f"{SUBJECTS_DIR}/freesurfer/{SUBJECT_ID}/label/rh.perirhinal_exvivo.thresh.label"
template_path = f"{SUBJECTS_DIR}/fmriprep/{SUBJECT_ID}/anat/{SUBJECT_ID}_desc-preproc_T1w.nii.gz"
# Define the output paths for the binary volume masks
output_path_left = f"{SUBJECTS_DIR}/freesurfer/{SUBJECT_ID}/mri/lh.perirhinal_exvivo.thresh.mask.nii.gz"
output_path_right = f"{SUBJECTS_DIR}/freesurfer/{SUBJECT_ID}/mri/rh.perirhinal_exvivo.thresh.mask.nii.gz"
# Convert the left hemisphere label to a binary volume mask
subprocess.run(["mri_label2vol", "--label", label_path_left, "--temp", template_path, "--subject", SUBJECT_ID, "--hemi", "lh", "--identity", "--fillthresh", "0.5", "--o", output_path_left], check=True)
# Convert the left hemisphere label to a binary volume mask
subprocess.run(["mri_label2vol", "--label", label_path_right, "--temp", template_path, "--subject", SUBJECT_ID, "--hemi", "rh", "--identity", "--fillthresh", "0.5", "--o", output_path_right], check=True)
from nilearn import plotting
plotting.plot_roi(output_path_left, bg_img=template_path, cmap="Set1", draw_cross=False)
plotting.plot_roi(output_path_right, bg_img=template_path, cmap="Set1", draw_cross=False)
however when I plot this mask, it doesn’t seem to be in the right place
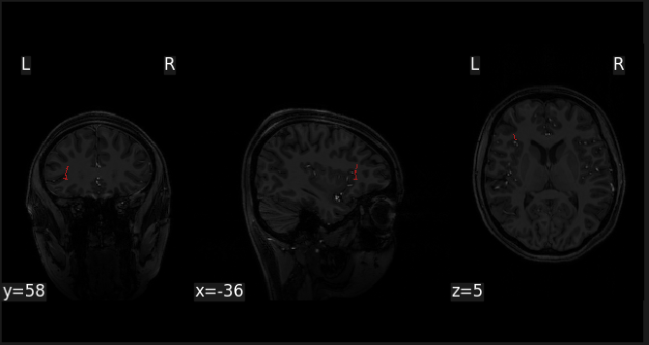
I’m guessing that I’m missing --reg regmatfile parameter, but I don’t know which file that would be…