Hello everyone,
I’m working on segmenting and visualizing the cerebral lobes from T1-weighted MRI scans using FreeSurfer. My objective is to:
- Segment the cerebral lobes from T1 MRI.
- Assign each lobe a different label and visualize them in 3D.
- Register the segmented lobes onto the original T1 scan for clear visualization.
I followed these steps in FreeSurfer:
Ran the full FreeSurfer pipeline:
recon-all -s my_subject -i T1.mgz -all
Converted the parcellation to individual labels:
mri_annotation2label --subject my_subject --hemi lh --annotation aparc.DKTatlas --outdir labels/
**Merged the labels into lobe-specific segmentations:
mri_mergelabels labels/lh.*.label -o lh_lobes.label
Converted the label files into volumetric images:
mri_label2vol --label lh_lobes.label --temp T1.mgz --o sujet1t1.nii --identity
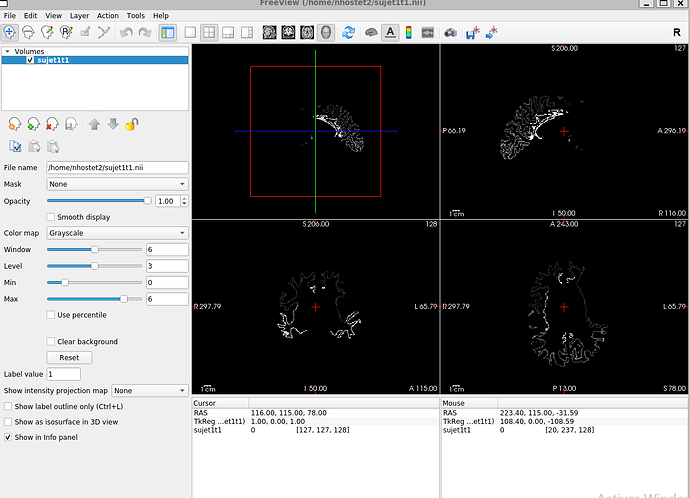
The Problem:
-
The output does not display as expected. Instead of solid, colored lobes, I see only contours outlining the regions
-
Applying
--apply_lutdoes not seem to fix the issue. -
I tried
mri_binarizeto ensure the labels were filled, but it didn’t change the visualization much.
Does anyone have experience with this type of segmentation? How can I ensure that each lobe is properly labeled, filled, and visualized in 3D correctly? Any help would be greatly appreciated!
Thank you in advance