Summary of what happened:
Hi all,
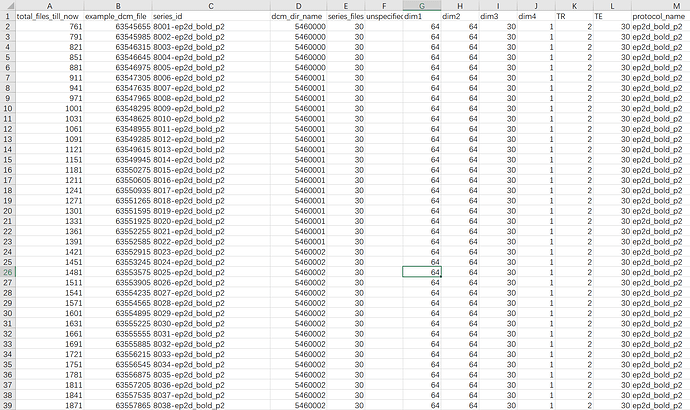
I checked the ‘.heudiconv’ folder, and ‘dicominfo.tsv’ showed my resting-state func scans have different ‘series_id’ (e.g., ‘8001-ep2d_bold_p2’,‘8002-ep2d_bold_p2’,…,‘8240-ep2d_bold_p2’)
Could I stack these 3d nii into 4d by Heudiconv?
Much appreciated
Command used (and if a helper script was used, a link to the helper script or the command generated):
- Command used in conda
heudiconv -d ./sourcedata/Site_CD5120/SS01/{subject}/DICOM/*/*/* -o ./rawdata/CD5120/SS01 -f ./sourcedata/heuristic_cd5120.py -s 009 010 011 012 013 014 015 016 017 018 -c dcm2niix -b --overwrite
- ‘heuristic_cd5120.py’
import os
def create_key(template, outtype=('nii.gz',), annotation_classes=None):
if template is None or not template:
raise ValueError('Template must be a valid format string')
return template, outtype, annotation_classes
def infotodict(seqinfo):
func = create_key('sub-{subject}/func/sub-{subject}_func_cd5120')
info = {anat: [], func: [], dti: []}
last_run = len(seqinfo)
for idx, s in enumerate(seqinfo):
if ('ep2d_bold' in s.protocol_name):
info[func].append(s.series_id)
return info
- ‘009.auto.txt’/‘009.edit.txt’ in ‘.heudiconv’:
{('sub-{subject}/func/sub-{subject}_func_cd5120', ('nii.gz',), None): ['8001-ep2d_bold_p2',
'8002-ep2d_bold_p2',
...
'8240-ep2d_bold_p2']}
- ‘filegroup.json’ in ‘.heudiconv’:
{
"8001-ep2d_bold_p2": [
"./sourcedata/Site_CD5120/SS01/009/DICOM/21070613/05460000/63545655",
"./sourcedata/Site_CD5120/SS01/009/DICOM/21070613/05460000/63545666",
...
"./sourcedata/Site_CD5120/SS01/009/DICOM/21070613/05460000/63546634"
],
...
"8240-ep2d_bold_p2": [
"./sourcedata/Site_CD5120/SS01/009/DICOM/21070613/05460015/63624525",
...
"./sourcedata/Site_CD5120/SS01/009/DICOM/21070613/05460015/63624844"
]
}
Version:
heudiconv version 0.13.1
dcm2niix version v1.0.20230411
Environment (Docker, Singularity, custom installation):
miniconda3 environment
Data format:
├── 009
│ └── DICOM
│ ├── 21070613
│ │ ├── 05460000
│ │ │ ├── 63541684
│ │ │ └── 63541695
│ │ │ └── ...
│ │ ├── 05460001
│ │ │ ├── 63547184
│ │ │ ├── 63547195
│ │ │ └── ...
Relevant log outputs:
INFO: Post-treating ./rawdata/CD5120/SS01/sub-009/func/sub-009_func_cd5120.json file
INFO: Converting ./rawdata/CD5120/SS01/sub-009/func/sub-009_func_cd5120 (30 DICOMs) -> ./rawdata/CD5120/SS01/sub-009/func . Converter: dcm2niix . Output types: ('nii.gz',)
230917-09:53:09,819 nipype.workflow INFO:
[Node] Setting-up "convert" in "/tmp/dcm2niix060ve0cm/convert".
INFO: [Node] Setting-up "convert" in "/tmp/dcm2niix060ve0cm/convert".
230917-09:53:09,843 nipype.workflow INFO:
[Node] Executing "convert" <nipype.interfaces.dcm2nii.Dcm2niix>
INFO: [Node] Executing "convert" <nipype.interfaces.dcm2nii.Dcm2niix>
230917-09:53:10,126 nipype.interface INFO:
stdout 2023-09-17T09:53:10.126776:Chris Rorden's dcm2niiX version v1.0.20230411 GCC12.2.0 x86-64 (64-bit Linux)
INFO: stdout 2023-09-17T09:53:10.126776:Chris Rorden's dcm2niiX version v1.0.20230411 GCC12.2.0 x86-64 (64-bit Linux)
230917-09:53:10,127 nipype.interface INFO:
stdout 2023-09-17T09:53:10.126776:Found 30 DICOM file(s)
INFO: stdout 2023-09-17T09:53:10.126776:Found 30 DICOM file(s)
230917-09:53:10,127 nipype.interface INFO:
stdout 2023-09-17T09:53:10.126776:Warning: Assuming mosaics saved in reverse order due to 'sSliceArray.ucImageNumb'
INFO: stdout 2023-09-17T09:53:10.126776:Warning: Assuming mosaics saved in reverse order due to 'sSliceArray.ucImageNumb'
230917-09:53:10,127 nipype.interface INFO:
stdout 2023-09-17T09:53:10.126776:Warning: Siemens XA exported as classic not enhanced DICOM (issue 236)
INFO: stdout 2023-09-17T09:53:10.126776:Warning: Siemens XA exported as classic not enhanced DICOM (issue 236)
230917-09:53:10,127 nipype.interface INFO:
stdout 2023-09-17T09:53:10.126776:Convert 30 DICOM as ./rawdata/CD5120/SS01/sub-009/func/sub-009_func_cd5120_heudiconv063 (64x64x30x1)
INFO: stdout 2023-09-17T09:53:10.126776:Convert 30 DICOM as ./rawdata/CD5120/SS01/sub-009/func/sub-009_func_cd5120_heudiconv063 (64x64x30x1)
230917-09:53:10,235 nipype.interface INFO:
stdout 2023-09-17T09:53:10.235645:Conversion required 0.351797 seconds (0.141253 for core code).
INFO: stdout 2023-09-17T09:53:10.235645:Conversion required 0.351797 seconds (0.141253 for core code).
230917-09:53:10,300 nipype.workflow INFO:
[Node] Finished "convert", elapsed time 0.455839s.
INFO: [Node] Finished "convert", elapsed time 0.455839s.
230917-09:53:10,394 nipype.workflow INFO:
[Node] Setting-up "embedder" in "/tmp/embedmetax0ey3pgs/embedder".
INFO: [Node] Setting-up "embedder" in "/tmp/embedmetax0ey3pgs/embedder".
230917-09:53:10,412 nipype.workflow INFO:
[Node] Executing "embedder" <nipype.interfaces.utility.wrappers.Function>
INFO: [Node] Executing "embedder" <nipype.interfaces.utility.wrappers.Function>
230917-09:53:11,244 nipype.workflow INFO:
[Node] Finished "embedder", elapsed time 0.828648s.
INFO: [Node] Finished "embedder", elapsed time 0.828648s.
230917-09:53:11,244 nipype.workflow WARNING:
Storing result file without outputs
WARNING: Storing result file without outputs
230917-09:53:11,245 nipype.workflow WARNING:
[Node] Error on "embedder" (/tmp/embedmetax0ey3pgs/embedder)
WARNING: [Node] Error on "embedder" (/tmp/embedmetax0ey3pgs/embedder)
ERROR: Embedding failed: Exception raised while executing Node embedder.
Traceback:
Traceback (most recent call last):
File "./miniconda3/envs/heudiconv/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "./miniconda3/envs/heudiconv/lib/python3.10/site-packages/nipype/interfaces/utility/wrappers.py", line 142, in _run_interface
out = function_handle(**args)
File "<string>", line 35, in embed_dicom_and_nifti_metadata
File "./miniconda3/envs/heudiconv/lib/python3.10/site-packages/dcmstack/dcmstack.py", line 1226, in parse_and_stack
results[key] = stack_group(group, warn_on_except, **stack_args)
File "./miniconda3/envs/heudiconv/lib/python3.10/site-packages/dcmstack/dcmstack.py", line 1180, in stack_group
result.add_dcm(dcm, meta)
File "./miniconda3/envs/heudiconv/lib/python3.10/site-packages/dcmstack/dcmstack.py", line 605, in add_dcm
nii_wrp = NiftiWrapper.from_dicom_wrapper(dw, meta)
File "./miniconda3/envs/heudiconv/lib/python3.10/site-packages/dcmstack/dcmmeta.py", line 1536, in from_dicom_wrapper
affine = np.dot(np.diag([-1., -1., 1., 1.]), dcm_wrp.get_affine())
AttributeError: 'Wrapper' object has no attribute 'get_affine'