Summary of what happened:
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run -B /zwork/hannah/baker/data:/bids-root
-B /zwork/hannah/baker/data/derivatives/aslprep:/output
-B /zwork/hannah/baker/workdir:/workdir
-B /sw/freesurfer/license.txt:/license
/zwork/hannah/baker/data/code/aslprep/aslprep-0.7.5.simg
/bids-root
/output
participant
--participant_label sub-01
-w /workdir
--fs-license-file /license
--longitudinal
--scorescrub
--basil
--skip_bids_validation
Version:
0.7.5
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity container on a shared computing cluster
Screenshots / relevant information:
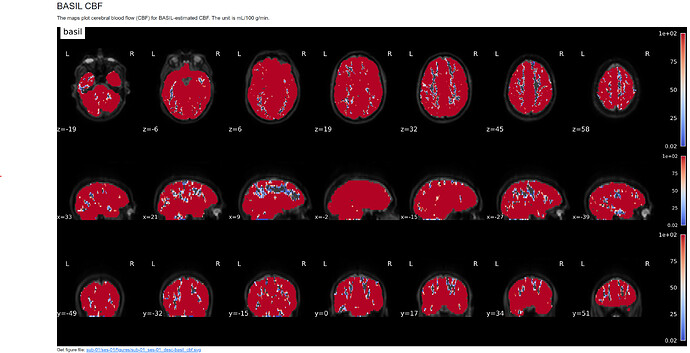
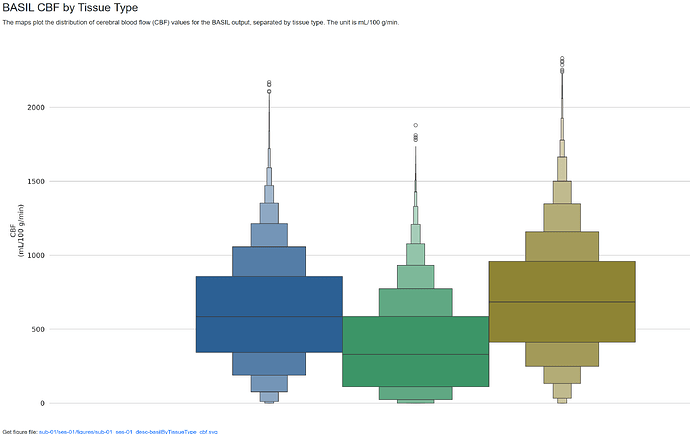
I’m working with a multi-PLD PCASL dataset from a collaborator using the ADNI4 protocol. I was able to successfully run ASLPrep without it crashing, but the CBF values are quite off (500+ mL/g/min, see attached images). This makes me think that the M0 image may not be scaled. I was given raw data for the ASL data but not the structural data so I ended up manually converting the data to BIDS using dcm2niix, therefore no scaling happened during preprocessing. I’m not sure if it’s relevant, but when I received the dicoms, there were two in each M0 folder and the sequence I’m used to working with only has 1 m0 dicom so I wasn’t sure if this might be part of the problem. Is there a way to find the value needed to scale M0 by, either in the MRI protocol or NIFTI header? I assume then I could use the --m0_scan flag and that might solve the issue. If it looks like it might be related to something else let me know. Thanks in advance!