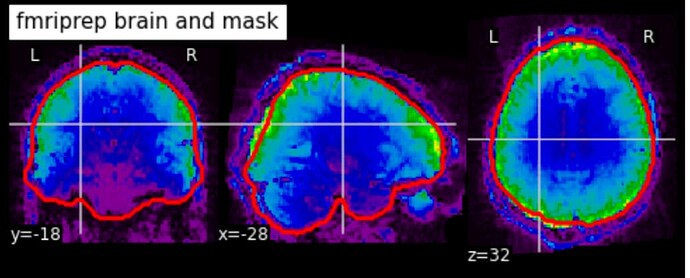
The brain mask generated by fMRIprep for my EPI is not bad but also not great (i.e. check out the back of the brain on the sagittal section, check out the dorsal part of the coronal section).
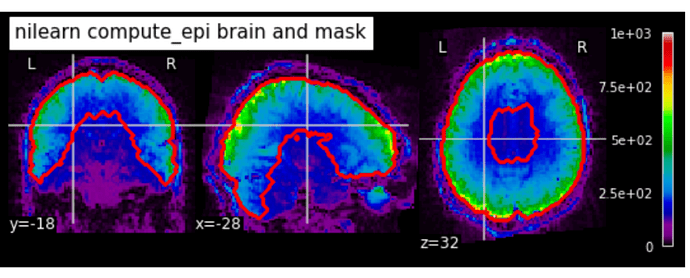
So I used nilearn’s compute_epi function. Interestingly, this function seems to hug the anatomy close for a better fit along the border of the cortex and fixes the aforementioned problems. However, it has the obvious issue of failing on the temporal lobes/ventral part. I tried changing the lower_cutoff value up and down, and it made zero difference. upper_cutoff also didn’t change anything.
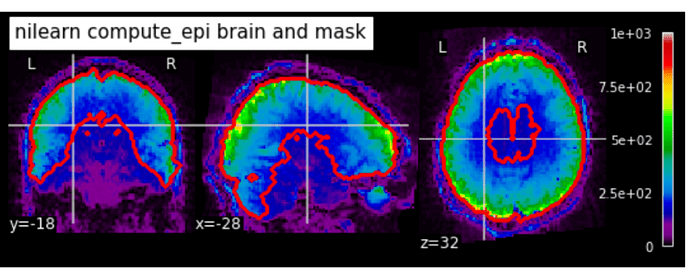
Changing ‘opening’ did have some interesting effects, but nothing good overall:
opening = 1
opening = False
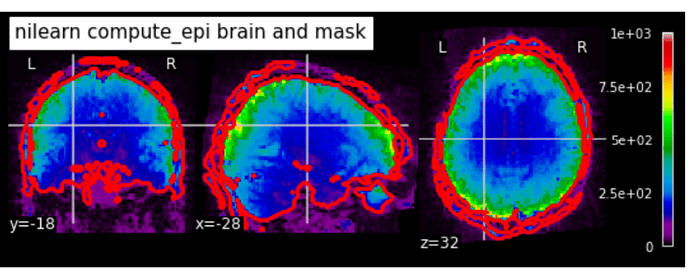
opening greater than 2 just made the problem worse.
any advice? is there anything i can tweak to get a good mask from this? am i overreacting about the fmriprep mask?
thank you ![]()