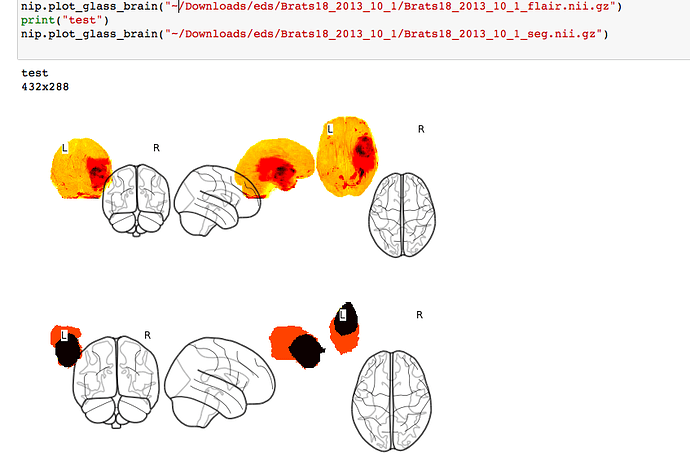
Hi. I’m attempting to plot nii images (from the BraTS dataset) using the plot_glass_brain function. However, the resultant image is all out of alignment. What can I do to fix this?
I’m thinking it could have something to do with those images not being in MNI space, so you may need to transform them. I’m not sure what the best way to transform the coordinates is but you could try this to see if it changes anything until a better suggesting comes along:
from nilearn import datasets, image
import nilearn.plotting as nip
img = image.load_img(‘path/to/your/file’)
mni = datasets.load_mni152_template()
img.affine = mni.affine.copy()
nip.plot_glass_brain(img)
Hi @Eli
Indeed, this is most likely due to the images not being in MNI space. You can resample your input image to the MNI152 template like this:
from nilearn.image import resample_to_img, load_img
from nilearn.datasets import load_mni152_template
mni = load_mni152_template()
niimg = load_img(path/to/img)
resampled = resample_to_img(niimg, mni)
Hope this helps!
For people looking to align their data, resample_to_img doesn’t, it merely resamples to the same voxel size as the target image.
From the documentation:
Resample a Niimg-like source image on a target Niimg-like image (no registration is performed: the image should already be aligned).
I’m still searching for a way to do alignment / registration…