Sorry in advance of the simplicity of this question, but I can’t find a solution anywhere.
Does anyone know the best way to visualize individual white matter tracts from templates such as natbrainlab or JHU? I have lesion maps and was able to identify a list of white matter tracts that pass through the lesion using BCBtoolkit tractotron. Now, I would like to overlay individual tracts on a t1 template in MNI space together with the lesion map to best visualize where specific tracts intersect the lesion and where else the WM tract travels. I would like to be able to display this using MRIcron multislice viewing tool as well as in a 3d glass brain using MRIcroGL, but I am open to other solutions. I’ve attached an image from Akinina et al., 2019 to demonstrate an example of what I am trying to do.
I know there must be a way to edit these white matter templates to select individual labels/colors to be visual or hidden/deleted, but I have not been able to figure this out. Any help would be much appreciated!
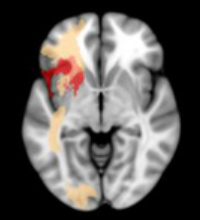
Link to full article from Akinina et al., 2019: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6650369/