If fmriprep is run twice on the same input dataset should I expect identical output images, or only very similar (e.g., due to randomness in parameter optimization)?
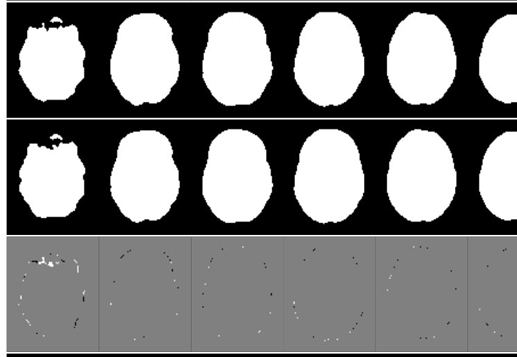
I am comparing the results of running fmriprep 1.3.2 twice on the the same dataset, with presumably identical input and settings. The confounds_regressors.tsv produced by the two runs are identical, but the images are not. For example, these are a few slices from the brain mask generated for the same participant and imaging run: first row is the first time fmriprep was run, second the second, and the third row is the difference between the two. The differences are clearly small and around the edges, but present.
Subtle differences are present in the functional images as well; not just in the edge voxels (as in this brain mask) but throughout the volume and in all frames. I haven’t compared the surface output yet.
The question: do you expect something on the order of this amount of variability between fmriprep runs, or do you expect identical output? If the output should be identical I will dig further into the settings/versions and conduct more tests to see what could be causing the discrepancy.
Thank you!