Dear Experts,
I successfully normalized EPI images to spm12\toolbox\OldNorm\EPI.nii.
Based on some literature, normalization should work also without anatomical images and the results look good.
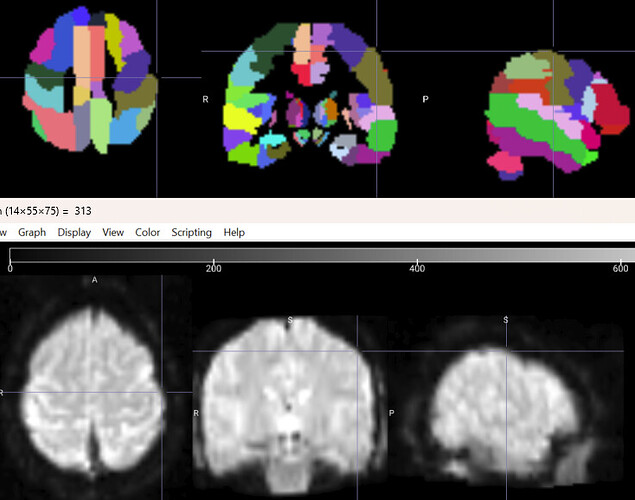
If I check the result with ‘MRIcro’ or the SPM tool ‘Check Reg’ by comparing it to an atlas, everything looks good:
However, the images are not exactly in the same space, especially since there is more empty space around the AAL3 atlas. Therefore, I cannot apply the masks to the data in a straightforward manner.
My question is probably rather simple: How do I now use Matlab/SPM (ideally in a scripted manner) to apply masks of the differen regions to the data?
Thank you!
Marco
1 Like
Thank you!
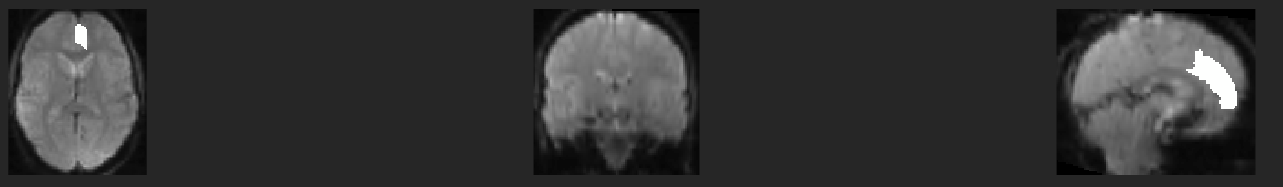
This script does not look like it is doing any sort of transformation. In this image I highlight Index 19 of AAL3 and it should end up at the border of the brain, not inside of it:
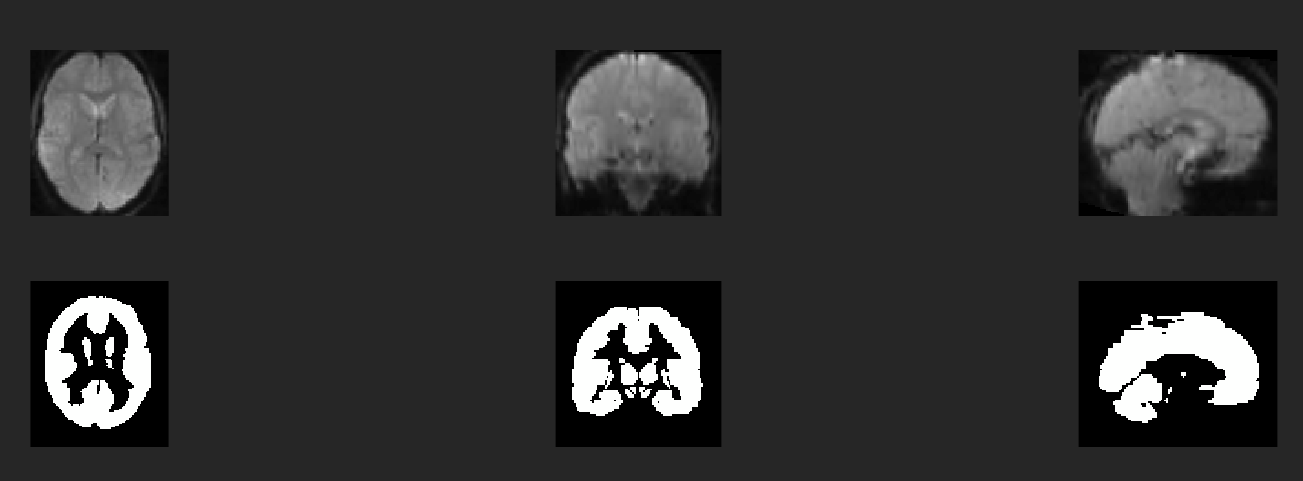
To again illustrate the problem again, here you see my data and the AAL3 atlas:
So you can see how much more empty space there is around the atlas than around spm12\toolbox\OldNorm\EPI.nii (which I used for normalization).
And I am looking for a way to apply the rois from the atlas to this data.
Hi Marco,
I had a different issue but maybe the approach is similar: Using ROIs created by SPM with fMRIPrep data
In brief, you could try using ANTs or FSL to compute registration parameters between the template that your atlas is based on and the template that you used to register you data to and then apply the resulting transformation parameters to your atlas-based ROIs. At least for my issue, this worked.
Best,
Jasmin
Hey @JasminStein,
thank you for your reply - I have ants running now and it works well it seems.
I think my issue is that I am lacking a “template that my atlas is based on”. If I compare for example \spm12\canonical\single_subj_T1.nii and the AAL3 atlas, they are both 91x109x91 in size, but there is significantly more ‘black space’ around the atlas regions.
I also asked the AAL-Team for an anatomical template for the AAL atlases that is in the exact same space now.
Best wishes,
Marco
1 Like
I see! I have used the SPM wfupickatlas tool which should create AAL ROIs in the space and size of the SPM template.