Hello,
I am trying to visualize a thresholded Z map (obtained from an ALE meta-analysis and corrected for multiple comparisons) that is based on the MNI152 space by converting it to surface space (fsLR). I would like to use the surfplot package for visualization, but I am encountering some issues along the way. Below, I describe my workflow and the problems I am facing.
What I did:
-
Converting the Z map to surface space:
from neuromaps import transforms control_all = '../results/control_all_z_size_level_thresh.nii' controls_surf = transforms.mni152_to_fslr(control_all, '32k') # Convert to fsLR 32k surface space- After the transformation, I obtained a result (
controls_surf) that contains data for both the left and right hemispheres.
- After the transformation, I obtained a result (
-
Saving the left and right hemisphere data as .gii files:
import nibabel as nib lh_data = controls_surf[0] # Left hemisphere data rh_data = controls_surf[1] # Right hemisphere data # Save the left and right hemisphere data to .gii files nib.save(lh_data, '../results/lh_data.gii') nib.save(rh_data, '../results/rh_data.gii') -
Loading the .gii files and visualizing with surfplot:
- I followed the tutorial from Surfplot Documentation to visualize
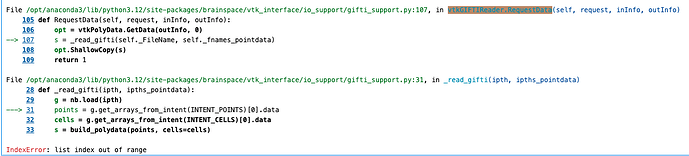
.giifiles. However, when running the code, I encountered the following error:from surfplot import Plot from nilearn.datasets import fetch_surf_fsaverage # Load the .gii files for left and right hemispheres lh_data = nib.load('../results/lh_data.gii') rh_data = nib.load('../results/rh_data.gii') # Load the fsLR surface template surfaces = fetch_surf_fsaverage() # Create a surfplot visualization p = Plot(lh_data, rh_data) - Error message:
- I followed the tutorial from Surfplot Documentation to visualize
ValueError: Surface must be a path-like string, an instance of BSPolyData, or None
I am running Mac OS. how can I resolve it?
Thanks for any advice.