Hi,
I want to get the cortical thickness of fsLR_den-91k space from the results of fmriprep. And I want to make this process consistent with what BOLD image did, i.e., “–cifti-output”. It appears that fmriprep use “wb_command -metric-resample” to resampe fsaverage to fsLR. But I want to know more details. So I wrote my code here, if anyone could confirm it or give me some suggestions?
First I resample the thickness file (i.e.,lh.thickness in Freesurfer output directory) from native space to fsaverage164k space, like this:
mri_surf2surf --srcsubject sub-F001 --trgsubject fsaverage --hemi lh --sval /home/z/DataStorage/XW/fMRIprep/fmriprep/freesurfer/sub-F001/surf/lh.thickness --tval /home/z/DataStorage/XW/fMRIprep/fmriprep/space-fsaverage_hemi-L.thickness.func.gii
After that, I had gotten the thickness file named space-fsaverage_hemi-L.thickness.func.gii in fsaverage space.
Second I want to resample this file to fsLR_32k space, like this:
wb_command -metric-resample space-fsaverage_hemi-L.thickness.func.gii $current_sphere_L $target_sphere_L ADAP_BARY_AREA space-fsLR32K_hemi-L.thickness.func.gii -area-metrics $current_area_L $target_area_L
I don’t run this code, because I could not confirm these necessary files including $current_sphere_L, $target_sphere_L, $current_area_L, and $target_area_L.
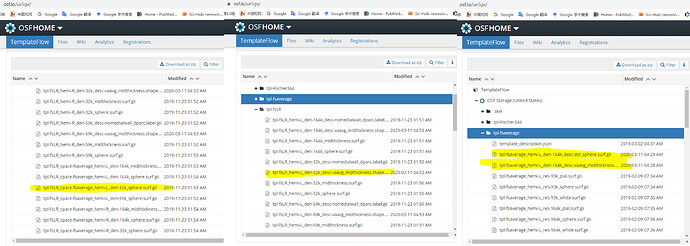
In my opinion, for $target_sphere_L , $target_area_L are files in fsLR32K, $current_sphere_L and $current_area_L, could I use the files in the yellow lines in the screenshot below?
Besides, are there other parameters that should be added in command “wb_command -metric-resample”? e.g., the flag [-largest].
Can someone give me some suggestions or know the specific commands of fmriprep?
Thanks a lot!