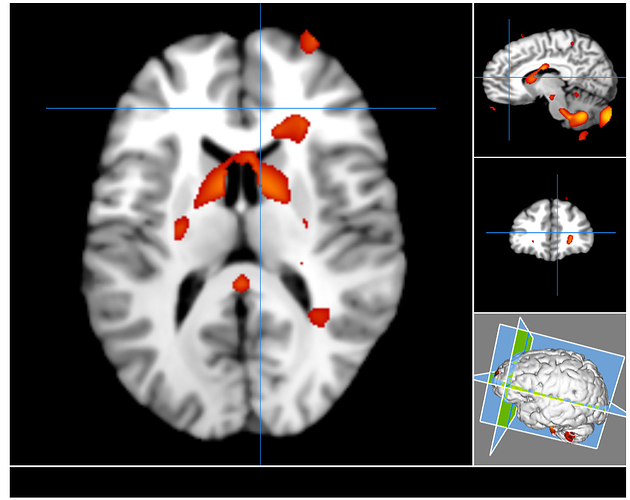
I want to perform a massive univariate analysis to compare structural grey matter images between two groups (patients and controls). I used nilearn’s NiftiMasker to smooth and standardize the data. After this, I used nilearn.mass_univariate.permuted_ols to perform voxel-wise contrasts between patients and controls. However, when I create a statistical image (t-values) based on the output from nilearn.mass_univariate.permuted_ols I get a very ‘blobby’ image with artifacts inside the ventricles and outside the brain:
I would like to check if a ‘classical’ SPM analysis reproduces the artifacts to rule out the possibility that the analysis technique causes these artifacts (to be honest, I am not sure if this should already be clear by the fact how nilearn.mass_univariate.permuted_ols works, or in other words, if it’s just not the right tool for this kind of analysis):
I really appreciate the possibility to perform SPM-analyses using Python and nipype, because I do not have access to Matlab on my PC and I also like the approach to have several analyses tools ‘under the hood’ using only Python. However, I don’t find good step-by-step tutorials to perform my analysis and I also don’t find the SPM syntax straightforward to read.
So my questions are:
1.) Is nilearn.mass_univariate.permuted_ols the right tool to perform my analysis or should I switch to SPM?
2.) Are there any step-by-step tutorials on how to use the nipype interface to perform SPM-analyses?
1 Like
Ahoi hoi @JohannesWiesner,
regarding your second question:
The SPM standalone version (compiled using MCR) might be of interest to you. You can either use it within a docker/singularity image (e.g., the nipype tutorial or the SPM BIDS App or install it on your system following the instructions provided here and call it within your nipype workflow as described here. For examples on how to use SPM within nipype please have a look here, here and here.
HTH, cheers, Peer
P.S.: maybe nistats would also be something for you? Here are examples of 2nd level analyses.
1.) Is nilearn.mass_univariate.permuted_ols the right tool to perform my analysis or should I switch to SPM?
Can you confirm you tried the steps from this example ?
In nistats, you may be particularly interested in non_parametric_inference in second level model module. But still it uses permuted_ols.
You should not use the permuted_ols function. It is meant for independent data, typically from second-level analyses.
You can use nistats for an SPM-similar Python suite.
Best,
Bertrand
Yes, I followed the steps from this example:
1.) Smoothing, standardizing and masking to MNI152 template (using NiftiMasker)
2.) Using permuted_ols to perform massive univariate analysis (with tested_vars = groups and target_vars = image data)
3.) using inverse_transform from niftimasker to create a t-value image based on the output t-values from permuted_ols
Can you explain why permuted_ols is used in the example but doesn’t work for my data set?