Dear Arno, dear all,
I was trying to run the function CurvatureMain from the Mindboggle Docker container, but the command was not found, neither “curvature”. Only “mindboggle” was found and seemed to partially work. However, I don’t need to run the whole workflow. Is it possible to run a single Mindboggle or FreeSurfer function in the Docker container, or do I have to install Mindboggle / FreeSurfer differently?
I have installed Docker on an Ubuntu 18.04.2 LTS virtual machine (Docker version 18.09.6, build 481bc77) and followed all the Mindboggle Docker installation instructions. If more details are needed please let me know.
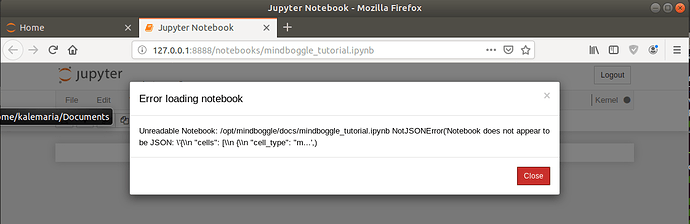
First, I wanted to try to find an answer in the Mindboggle jupyter notebook tutorial. However, when I tried to run jupyter notebook from the Docker container as described in “Getting help” on the Mindboggle (github) page, an error occurred:
OSError: [Errno 99] Cannot assign requested address
I found here and here (issue 168) that one has to call jupyter notebook with options “–ip=0.0.0.0 –port=8080 --allow-root”, then jupyter notebook launched in the terminal, but not in a browser, and a NotebookApp error occurred:
No web browser found: could not locate runnable browser.
Copying the URL with the token to a browser did not work. Also “port=8888” and “–ip=127.0.0.1” (as advised here) and different combinations of the options did not help.
The issue 168 with a related problem to mine has latest answer: “We forgot to update this doc after switching to neurodocker. The command mindboggle123 has to be specified after the docker image name.” But I could not understand where one is supposed to specify “mindboggle123”.
I am looking forward to any help. Thanks in advance!
Ria
 I’ll try it again.
I’ll try it again.