I would like to replicate the fmriprep compcor workflow to result in confounding timeseries .tsv and metadata .json. Here is what I am trying to refer to create a script in my local machine which should output what fmriprep outputs from compcor workflow
I tried implementing point 4 from above points and I was able to generate confounds timeseries .tsv and metadata .json but I noticed in the github code and fmriprep doc(point 2) that there is lot of preprocessing steps done for mask_files (cbf, wm, combined) to pass in as parameter in compcor Node. So here are my questions
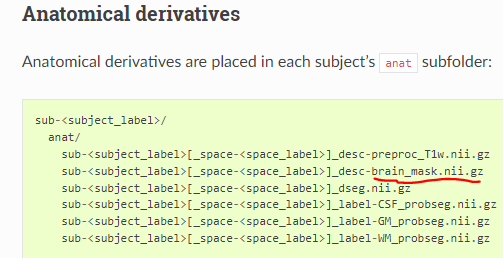
- Where can I find the mask_files(CSF, WM, combined) to add in compcor package? so that I will pass mask_names which will print into metadata
- How can I run compcor in my local machine to match the outputs of fmriprep confounds?
- Is there any reference I can follow to implement this functionality?
- is the highlighted file union(combined mask) of wm_probseg mask and csf_probseg mask?
It would be great for my learning if I can get some guidance on this concept
thanks