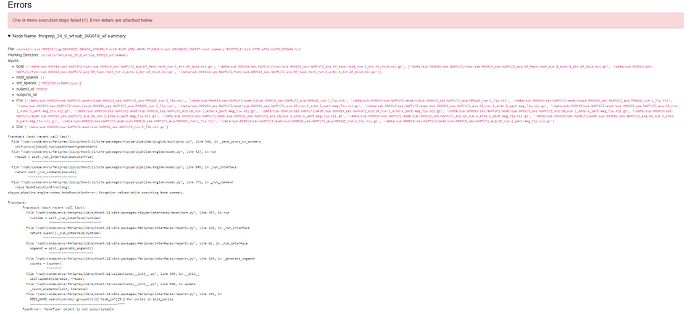
Summary of what happened: I am new to both MRI and fMRIprep and I am trying to use it to preprocess a dataset and keep running into errors on the BIDS format and other errors. Colleagues have mentioned running into similar issues due to the multi echo and multi channel data we are using. here is the latest error code I get from the generated html file:
Command used (and if a helper script was used, a link to the helper script or the command generated): source fmriprep.sh
PASTE CODE HERE
#!/bin/bash
#User inputs:
bids_root_dir=/home/trepie/prevent_rsfMRI
subj=999919
nthreads=16
mem=40 #gb
container=docker docker or singularity
#Begin:
#Convert virtual memory from gb to mb
mem=echo "${mem//[!0-9]/}" #remove gb at end
mem_mb=echo $(((mem*1000)-5000)) #reduce some memory for buffer space during pre-processing
export TEMPLATEFLOW_HOME=$HOME/.cache/templateflow
export FS_LICENSE=$HOME/Downloads/license.txt
#Run fmriprep
if [ $container == singularity ]; then
unset PYTHONPATH; singularity run -B $HOME/.cache/templateflow:/opt/templateflow $HOME/fmriprep.simg
$bids_root_dir $bids_root_dir/derivatives
participant
–participant-label $subj
–skip-bids-validation
–md-only-boilerplate
–fs-license-file $FREESURFER_HOME/license.txt
–fs-no-reconall
–output-spaces MNI152NLin2009cAsym:res-2
–nthreads $nthreads
–stop-on-first-crash
–mem_mb $mem_mb
-w $HOME
else
fmriprep-docker $bids_root_dir $bids_root_dir/derivatives
participant
–participant-label $subj
-md-only-boilerplate
–fs-license-file $HOME/Downloads/license.txt
–fs-no-reconall
–output-spaces MNI152NLin2009cAsym:res-2
–nthreads $nthreads
–stop-on-first-crash
–mem_mb $mem_mb
-w $HOME
fi
Version: v23.2.3
Environment (Docker, Singularity / Apptainer, custom installation): Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
bids-validator@1.14.6
(node:9) Warning: Closing directory handle on garbage collection
(Use node --trace-warnings ... to show where the warning was created)
1: [ERR] Files with such naming scheme are not part of BIDS specification. This error is most commonly caused by typos in file names that make them not BIDS compatible. Please consult the specification and make sure your files are named correctly. If this is not a file naming issue (for example when including files not yet covered by the BIDS specification) you should include a “.bidsignore” file in your dataset (see GitHub - bids-standard/bids-validator: Validator for the Brain Imaging Data Structure for details). Please note that derived (processed) data should be placed in /derivatives folder and source data (such as DICOMS or behavioural logs in proprietary formats) should be placed in the /sourcedata folder. (code: 1 - NOT_INCLUDED)
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-1_b1map.json
Evidence: sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-1_b1map.json
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-1_b1map.nii.gz
Evidence: sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-1_b1map.nii.gz
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-2_b1map.json
Evidence: sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-2_b1map.json
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-2_b1map.nii.gz
Evidence: sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-2_b1map.nii.gz
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-1_b1map.json
Evidence: sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-1_b1map.json
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-1_b1map.nii.gz
Evidence: sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-1_b1map.nii.gz
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-2_b1map.json
Evidence: sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-2_b1map.json
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-2_b1map.nii.gz
Evidence: sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-2_b1map.nii.gz
./sub-999919/ses-NAPFU72/MTL0203_999919_NAPFU72_ep2d_diff_acc6-AP_b0_ep2d_diff_acc6-AP_b0_TE1.bval
Evidence: MTL0203_999919_NAPFU72_ep2d_diff_acc6-AP_b0_ep2d_diff_acc6-AP_b0_TE1.bval
./sub-999919/ses-NAPFU72/MTL0203_999919_NAPFU72_ep2d_diff_acc6-AP_b0_ep2d_diff_acc6-AP_b0_TE1.bvec
Evidence: MTL0203_999919_NAPFU72_ep2d_diff_acc6-AP_b0_ep2d_diff_acc6-AP_b0_TE1.bvec
… and 745 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=NOT_INCLUDED for existing conversations about this issue.
2: [ERR] acq Name contain an Illegal Character hyphen or underscore. Please edit the filename as per BIDS spec. (code: 59 - ACQ_NAME_CONTAIN_ILLEGAL_CHARACTER)
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-1_b1map.json
Evidence: acq name contains illegal character:/sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-1_b1map.json
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-2_b1map.json
Evidence: acq name contains illegal character:/sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_Siemens_acq-_ep_run-2_b1map.json
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-1_b1map.json
Evidence: acq name contains illegal character:/sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-1_b1map.json
./sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-2_b1map.json
Evidence: acq name contains illegal character:/sub-999919/ses-NAPFU72/B1map/sub-999919_ses-NAPFU72_acq-_ep_acq-_ep_run-2_b1map.json
Please visit https://neurostars.org/search?q=ACQ_NAME_CONTAIN_ILLEGAL_CHARACTER for existing conversations about this issue.
3: [ERR] Ses label contain an Illegal Character hyphen or underscore. Please edit the filename as per BIDS spec. (code: 63 - SESSION_VALUE_CONTAINS_ILLEGAL_CHARACTER)
./sub-999919/ses-NAPFU72/anat/QSM/corrected_phase/src/sub-999919_ses-NAPFU72_mag_1_mean_N4_bet.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/QSM/corrected_phase/src/sub-999919_ses-NAPFU72_mag_1_mean_N4_bet.nii.gz
./sub-999919/ses-NAPFU72/anat/QSM/corrected_phase/src/sub-999919_ses-NAPFU72_mag_1_mean_N4_bet_corr_inT1w_space.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/QSM/corrected_phase/src/sub-999919_ses-NAPFU72_mag_1_mean_N4_bet_corr_inT1w_space.nii.gz
./sub-999919/ses-NAPFU72/anat/QSM/corrected_phase/src/sub-999919_ses-NAPFU72_mag_1_mean_N4_bet_mask.nii
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/QSM/corrected_phase/src/sub-999919_ses-NAPFU72_mag_1_mean_N4_bet_mask.nii
./sub-999919/ses-NAPFU72/anat/QSM/sub-999919_ses-NAPFU72_qsm_space-mni.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/QSM/sub-999919_ses-NAPFU72_qsm_space-mni.nii.gz
./sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-dia.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-dia.nii.gz
./sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-dir.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-dir.nii.gz
./sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-label.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-label.nii.gz
./sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-proba.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-proba.nii.gz
./sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-pv_space-mni.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-pv_space-mni.nii.gz
./sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-seg.nii.gz
Evidence: ses name contains illegal character:/sub-999919/ses-NAPFU72/anat/veins/sub-999919_ses-NAPFU72_qsm_mvf-seg.nii.gz
... and 14 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=SESSION_VALUE_CONTAINS_ILLEGAL_CHARACTER for existing conversations about this issue.
4: [ERR] NIfTI file exist with both '.nii' and '.nii.gz' extensions. (code: 74 - DUPLICATE_NIFTI_FILES)
./sub-999919/ses-NAPFU72/anat/sub-999919_ses-NAPFU72_acq-MPRAGE_run-1_T1w.nii
./sub-999919/ses-NAPFU72/anat/sub-999919_ses-NAPFU72_acq-MPRAGE_run-1_T1w.nii.gz
Please visit https://neurostars.org/search?q=DUPLICATE_NIFTI_FILES for existing conversations about this issue.
1: [WARN] The recommended file /README is missing. See Section 03 (Modality agnostic files) of the BIDS specification. (code: 101 - README_FILE_MISSING)
Please visit https://neurostars.org/search?q=README_FILE_MISSING for existing conversations about this issue.
Summary: Available Tasks: Available Modalities:
970 Files, 4.03GB MRI
2 - Subjects
2 - Sessions
If you have any questions, please post on https://neurostars.org/tags/bids.
Traceback (most recent call last):
File “/opt/conda/envs/fmriprep/bin/fmriprep”, line 8, in
sys.exit(main())
^^^^^^
File “/opt/conda/envs/fmriprep/lib/python3.11/site-packages/fmriprep/cli/run.py”, line 40, in main
parse_args()
File “/opt/conda/envs/fmriprep/lib/python3.11/site-packages/fmriprep/cli/parser.py”, line 903, in parse_args
validate_input_dir(
File “/opt/conda/envs/fmriprep/lib/python3.11/site-packages/fmriprep/utils/bids.py”, line 245, in validate_input_dir
subprocess.check_call([‘bids-validator’, str(bids_dir), ‘-c’, temp.name]) # noqa: S607
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File “/opt/conda/envs/fmriprep/lib/python3.11/subprocess.py”, line 413, in check_call
raise CalledProcessError(retcode, cmd)
subprocess.CalledProcessError: Command ‘[‘bids-validator’, ‘/data’, ‘-c’, ‘/tmp/tmpb51y6vxe.json’]’ returned non-zero exit status 1.
fMRIPrep: Please report errors to Issues · nipreps/fmriprep · GitHub
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE