I am frustrated. I want to run my standalone application of .m scripts using spm12 on linux server. Is there any easier way to tackle first error about ‘cfg_getfile’
Which option from spm_make_standalone are you referring to? The compiler is configured to include all SPM files.
If you only one to execute scripts (i.e. not defining new functions) then you don’t need to compile yourself and can use the standalone SPM linked above.
I need SPM12 with MCR 2018b. Let me try to compile by your method then I’ll let you know. Thanks Alot
OK, let me know how it goes. If you give me a bit of time, I could compile one version for you with R2018b. Searching through old files, I could find this (r7487, R2018b).
I also have used this r7487, and thay cfg_getfile error was using this one. And yes if you can compile one version for me I’ll be very very thankful to you. I am using R2018b MCR and Matlab for Linux
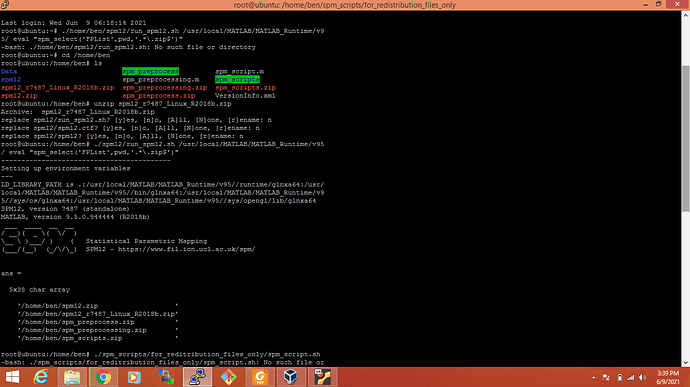
I am surprised you got the cfg_getfile error with that version. I tried installing and using it myself and it seems to work fine:
wget https://www.fil.ion.ucl.ac.uk/spm/download/restricted/utopia/dev/soon_gone/spm12_r7487_Linux_R2018b.zip
wget https://ssd.mathworks.com/supportfiles/downloads/R2018b/deployment_files/R2018b/installers/glnxa64/MCR_R2018b_glnxa64_installer.zip
unzip MCR_R2018b_glnxa64_installer.zip
sudo ./install -mode silent -agreeToLicense yes
unzip spm12_r7487_Linux_R2018b.zip
./spm12/run_spm12.sh /usr/local/MATLAB/MATLAB_Runtime/v95/ eval "spm_select('FPList',pwd,'.*\.zip$')"
which will display:
SPM12, version 7487 (standalone)
MATLAB, version 9.5.0.944444 (R2018b)
___ ____ __ __
/ __)( _ \( \/ )
\__ \ )___/ ) ( Statistical Parametric Mapping
(___/(__) (_/\/\_) SPM12 - https://www.fil.ion.ucl.ac.uk/spm/
ans =
2x58 char array
'/tmp/R2018b/MCR_R2018b_glnxa64_installer.zip'
'/tmp/R2018b/spm12_r7487_Linux_R2018b.zip '
Doest this work for you?
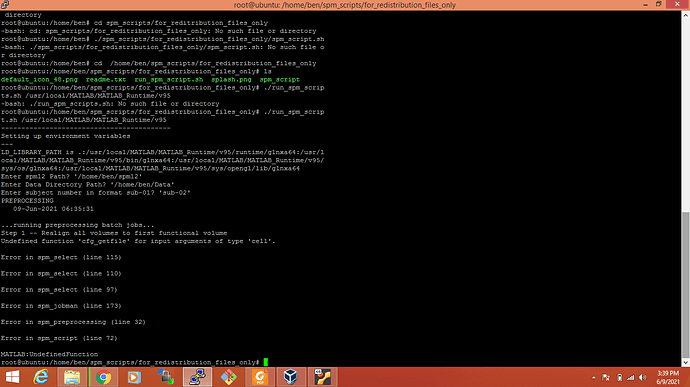
Worked. SPM is working. But while executing the shell script of my code that I want to run on Linux server. The same
code is working properly on windows and Linux DesktopProblem is same
I think you need to be a bit more explicit of what you are doing with your scripts. From what I see on the screenshots, it looks like you are trying to compile them? It seems that you have two *.m files, spm_script.m and spm_preprocessing.m: are they scripts or functions (starting with function spm_*)? Best would be to have a single script, that therefore does not need to be compiled and that you can directly provide to the SPM standalone command line:
./spm12/run_spm12.sh /usr/local/MATLAB/MATLAB_Runtime/v95/ script spm_script.m
How can I share my scripts with you? It’s not allowing to upload .m files. Actually spm_script is name given by me and spm_preprocessing also. But inside there is customized code to preprocess data. Spm_precosessing function is called in spm_script and it takes 4 parameters one for spm12 directory, second functional image, third analytical image and fourth cell size.
SPM help is usually through its discussion list or directly via email at fil.spm@ucl.ac.uk.
To upload your files here, perhaps rename them as *.txt beforehand.
%% MATLAB AND SPM12 BATCH SCRIPTING EXAMPLE:
% Processing a task-based single-subject fMRI dataset.
%
% Prerequisites:
% - Matlab recent version (developed using 2016b on Mac OS Sierra)
% - SPM12 neuroimage processing toolbox: SPM12 Software - Statistical Parametric Mapping
% - Task fMRI data: OpenNeuro
% - A basic understanding of the processing steps involved in analysing
% task fMRI
%
% Dependencies:
% - spm_preprocessing
%% INITIALIZATION
% Initialize file locations, image file names and other variables
if (isdeployed==true)
addpath(’/home/ben/spm12’);
end
prompt = 'Enter spm12 Path? ';
spm_dir = input(prompt);
%spm_dir = ‘/home/ben/spm12’;
prompt = 'Enter Data Directory Path? ';
data_dir = input(prompt);
%data_dir = ‘/home/ben/Data’;
addpath(spm_dir);
prompt = 'Enter subject number in format sub-01? ';
subj = input(prompt);
%subj = ‘sub-01’;
s_fn = [data_dir filesep subj filesep ‘anat’ filesep subj ‘_T1w.nii’];
f_fn = [data_dir filesep subj filesep ‘func’ filesep subj ‘_task-passiveimageviewing_bold.nii’];
stats_dir = [data_dir filesep subj filesep ‘stats’];
if ~exist(stats_dir,‘dir’)
mkdir(stats_dir)
end
fwhm = 6; % mm
%% PREPROCESSING
disp(‘PREPROCESSING’)
disp(datetime(‘now’));
%t1 = datetime(‘now’);
% Preprocess structural and functional images (if not already)
[d, f, e] = fileparts(s_fn);
[d1, f1, e1] = fileparts(f_fn);
if exist([d filesep ‘rc1’ f e], ‘file’)
disp(’…preproc already done, saving variables…’)
preproc_data = struct;
% Structural filenames
preproc_data.forward_transformation = [d filesep ‘y_’ f e];
preproc_data.inverse_transformation = [d filesep ‘iy_’ f e];
preproc_data.gm_fn = [d filesep ‘c1’ f e];
preproc_data.wm_fn = [d filesep ‘c2’ f e];
preproc_data.csf_fn = [d filesep ‘c3’ f e];
preproc_data.bone_fn = [d filesep ‘c4’ f e];
preproc_data.soft_fn = [d filesep ‘c5’ f e];
preproc_data.air_fn = [d filesep ‘c6’ f e];
preproc_data.rstructural_fn = [d filesep ‘r’ f e];
preproc_data.rgm_fn = [d filesep ‘rc1’ f e];
preproc_data.rwm_fn = [d filesep ‘rc2’ f e];
preproc_data.rcsf_fn = [d filesep ‘rc3’ f e];
preproc_data.rbone_fn = [d filesep ‘rc4’ f e];
preproc_data.rsoft_fn = [d filesep ‘rc5’ f e];
preproc_data.rair_fn = [d filesep ‘rc6’ f e];
% Functional filenames
preproc_data.rfunctional_fn = [d1 filesep ‘r’ f1 e1];
preproc_data.srfunctional_fn = [d1 filesep ‘sr’ f1 e1];
preproc_data.mp_fn = [d1 filesep ‘rp_’ f1 ‘.txt’];
preproc_data.MP = load(preproc_data.mp_fn);
else
disp(’…running preprocessing batch jobs…’)
preproc_data = spm_preprocessing(f_fn, s_fn, fwhm, spm_dir);
end
% Check coregistration and segmentation
spm_check_registration(s_fn, [preproc_data.rfunctional_fn ‘,1’], preproc_data.rgm_fn, preproc_data.rwm_fn, preproc_data.rcsf_fn)
disp(‘Preprocessing done!’)
disp(datetime(‘now’));
%t2 = datetime(‘now’);
function output = spm_preprocessing(functional4D_fn, structural_fn, fwhm, spm_dir)
% Function to complete preprocessing of structural and functional data from
% a single subject for use in any other Matlab/SPM12 script.
% Steps include coregistering structural image to first functional image,
% segmenting the coregistered structural image into tissue types, and
% reslicing the segments to the functional resolution image grid.
% Makes use of spm12 batch routines. If spm12 batch parameters are not
% explicitly set, defaults are assumed.
%
% INPUT:
% funcional4D_fn - filename of pre-real-time functional scan
% structural_fn - filename of T1-weighted structural scan
% fwhm - kernel size for smoothing operations
% spm_dir - SPM12 directory location
%
% OUTPUT:
% output - structure with filenames and data
%__________________________________________________________________________
%
% Load data
f4D_spm = spm_vol(functional4D_fn);
spm_size = size(f4D_spm);
Nt = spm_size(1);
% Declare output structure
output = struct;
% STEP 1 – Realign (estimate and reslice) all functionals to first functional
disp(‘Step 1 – Realign all volumes to first functional volume’);
spm(‘defaults’,‘fmri’);
spm_jobman(‘initcfg’);
realign_estimate_reslice = struct;
% Data
fnms={};
for i = 1:Nt
fnms{i} = [functional4D_fn ‘,’ num2str(i) ];
end
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.data={fnms’};
% Eoptions
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.eoptions.quality = 0.9;
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.eoptions.sep = 4;
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.eoptions.fwhm = 5;
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.eoptions.rtm = 1;
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.eoptions.interp = 2;
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.eoptions.wrap = [0 0 0];
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.eoptions.weight = ‘’;
% Roptions
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.roptions.which = [2 1];
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.roptions.interp = 4;
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.roptions.wrap = [0 0 0];
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.roptions.mask = 1;
realign_estimate_reslice.matlabbatch{1}.spm.spatial.realign.estwrite.roptions.prefix = ‘r’;
% Run
spm_jobman(‘run’,realign_estimate_reslice.matlabbatch);
[d, f, e] = fileparts(functional4D_fn);
output.rfunctional_fn = [d filesep ‘r’ f e];
output.mp_fn = [d filesep ‘rp_’ f ‘.txt’];
output.MP = load(output.mp_fn);
disp(‘Step 1 - Done!’);
% STEP 2 – Coregister structural image to first dynamic image (estimate only)
disp(‘Step 2 – Coregister structural image to first dynamic image’);
spm(‘defaults’,‘fmri’);
spm_jobman(‘initcfg’);
coreg_estimate = struct;
% Ref
coreg_estimate.matlabbatch{1}.spm.spatial.coreg.estimate.ref = {[functional4D_fn ‘,1’]};
% Source
coreg_estimate.matlabbatch{1}.spm.spatial.coreg.estimate.source = {structural_fn};
% Eoptions
coreg_estimate.matlabbatch{1}.spm.spatial.coreg.estimate.eoptions.cost_fun = ‘nmi’;
coreg_estimate.matlabbatch{1}.spm.spatial.coreg.estimate.eoptions.sep = [4 2];
coreg_estimate.matlabbatch{1}.spm.spatial.coreg.estimate.eoptions.tol = [0.02 0.02 0.02 0.001 0.001 0.001 0.01 0.01 0.01 0.001 0.001 0.001];
coreg_estimate.matlabbatch{1}.spm.spatial.coreg.estimate.eoptions.fwhm = [7 7];
% Run
spm_jobman(‘run’,coreg_estimate.matlabbatch);
disp(‘Step 2 - Done!’);
% STEP 3 – Segmentation of coregistered structural image into GM, WM, CSF, etc
% (with implicit warping to MNI space, saving forward and inverse transformations)
disp(‘Step 3 – Segmentation’);
spm(‘defaults’,‘fmri’);
spm_jobman(‘initcfg’);
segmentation = struct;
% Channel
segmentation.matlabbatch{1}.spm.spatial.preproc.channel.biasreg = 0.001;
segmentation.matlabbatch{1}.spm.spatial.preproc.channel.biasfwhm = 60;
segmentation.matlabbatch{1}.spm.spatial.preproc.channel.write = [0 1];
segmentation.matlabbatch{1}.spm.spatial.preproc.channel.vols = {structural_fn};
% Tissue
for t = 1:6
segmentation.matlabbatch{1}.spm.spatial.preproc.tissue(t).tpm = {[spm_dir filesep ‘tpm’ filesep ‘TPM.nii,’ num2str(t)]};
segmentation.matlabbatch{1}.spm.spatial.preproc.tissue(t).ngaus = t-1;
segmentation.matlabbatch{1}.spm.spatial.preproc.tissue(t).native = [1 0];
segmentation.matlabbatch{1}.spm.spatial.preproc.tissue(t).warped = [0 0];
end
segmentation.matlabbatch{1}.spm.spatial.preproc.tissue(1).ngaus = 1;
segmentation.matlabbatch{1}.spm.spatial.preproc.tissue(6).ngaus = 2;
% Warp
segmentation.matlabbatch{1}.spm.spatial.preproc.warp.mrf = 1;
segmentation.matlabbatch{1}.spm.spatial.preproc.warp.cleanup = 1;
segmentation.matlabbatch{1}.spm.spatial.preproc.warp.reg = [0 0.001 0.5 0.05 0.2];
segmentation.matlabbatch{1}.spm.spatial.preproc.warp.affreg = ‘mni’;
segmentation.matlabbatch{1}.spm.spatial.preproc.warp.fwhm = 0;
segmentation.matlabbatch{1}.spm.spatial.preproc.warp.samp = 3;
segmentation.matlabbatch{1}.spm.spatial.preproc.warp.write=[1 1];
% Run
spm_jobman(‘run’,segmentation.matlabbatch);
% Save filenames
[d, f, e] = fileparts(structural_fn);
output.forward_transformation = [d filesep ‘y_’ f e];
output.inverse_transformation = [d filesep ‘iy_’ f e];
output.gm_fn = [d filesep ‘c1’ f e];
output.wm_fn = [d filesep ‘c2’ f e];
output.csf_fn = [d filesep ‘c3’ f e];
output.bone_fn = [d filesep ‘c4’ f e];
output.soft_fn = [d filesep ‘c5’ f e];
output.air_fn = [d filesep ‘c6’ f e];
disp(‘Step 3 - done!’);
% STEP 4 – Reslice all to functional-resolution image grid
disp(‘Step 4 – Reslice all to functional-resolution image grid’);
spm(‘defaults’,‘fmri’);
spm_jobman(‘initcfg’);
reslice = struct;
% Ref
reslice.matlabbatch{1}.spm.spatial.coreg.write.ref = {[functional4D_fn ‘,1’]};
% Source
source_fns = {};
for i = 1:6
source_fns{i} = [d filesep ‘c’ num2str(i) f e];
end
source_fns{7} = structural_fn;
reslice.matlabbatch{1}.spm.spatial.coreg.write.source = source_fns’;
% Roptions
reslice.matlabbatch{1}.spm.spatial.coreg.write.roptions.interp = 4;
reslice.matlabbatch{1}.spm.spatial.coreg.write.roptions.wrap = [0 0 0];
reslice.matlabbatch{1}.spm.spatial.coreg.write.roptions.mask = 0;
reslice.matlabbatch{1}.spm.spatial.coreg.write.roptions.prefix = ‘r’;
% Run
spm_jobman(‘run’,reslice.matlabbatch);
% Save filenames
[d, f, e] = fileparts(structural_fn);
output.rstructural_fn = [d filesep ‘r’ f e];
output.rgm_fn = [d filesep ‘rc1’ f e];
output.rwm_fn = [d filesep ‘rc2’ f e];
output.rcsf_fn = [d filesep ‘rc3’ f e];
output.rbone_fn = [d filesep ‘rc4’ f e];
output.rsoft_fn = [d filesep ‘rc5’ f e];
output.rair_fn = [d filesep ‘rc6’ f e];
disp(‘Step 4 - done!’);
% STEP 5 – Gaussian kernel smoothing of realigned data
disp(‘STEP 5 – Gaussian kernel smoothing of realigned data’);
spm(‘defaults’,‘fmri’);
spm_jobman(‘initcfg’);
smooth = struct;
% Data
fns={};
for i = 1:Nt
fns{i} = [output.rfunctional_fn ‘,’ num2str(i) ];
end
smooth.matlabbatch{1}.spm.spatial.smooth.data = fns’;
% Other
smooth.matlabbatch{1}.spm.spatial.smooth.fwhm = [fwhm fwhm fwhm];
smooth.matlabbatch{1}.spm.spatial.smooth.dtype = 0;
smooth.matlabbatch{1}.spm.spatial.smooth.im = 0;
smooth.matlabbatch{1}.spm.spatial.smooth.prefix = ‘s’;
% Run
spm_jobman(‘run’,smooth.matlabbatch);
[d, f, e] = fileparts(output.rfunctional_fn);
output.srfunctional_fn = [d filesep ‘s’ f e];
disp(‘Step 5 - done!’);
This is the code that I am using. First one script that calls other files inside of it. This code is working properly on Windows and Linux Desktop.
How do you run it under Windows and Linux Desktop and how with the standalone version?
If you are using the standalone spm, it may not accept the function keyword in your script.
These two codes I have shared are my .m scripts. First one I just run on Matlab windows and also in Desktop Linux it Automatically calls other files for preprocessing.
And for Linux server I have created standalone of first script and added second one as required files and run its shell script by enter path of spm12 standalone. It’s showing errors.
So the working scripts are running inside matlab in both windows and linux, but the issue is with the compiled standalone spm right?
I suggest you start from a hello word one line .m script and run that with the standalone version. If that works, the go ahead step by step to identify the cause of the errors.
So you have one script spm_script.m and one function spm_preprocessing.m that you want to use with SPM standalone. A function has to be compiled beforehand in order to be usable in a standalone SPM. You therefore have two options:
- Compile your own SPM standalone that will include all of SPM and
spm_preprocessing.m. To do so, copyspm_preprocessing.min your SPM installation and runspm_make_standalone. From that compiled version, you can run your script (that will call internallyspm_preprocessing.m) with:
./run_spm12.sh /usr/local/MATLAB/MATLAB_Runtime/v95/ script spm_script.m
- Use SPM standalone as is (btw, see this for R2018b) and make sure to not define a new function. In your case, you would copy the content of
spm_preprocessing.m(without the first linefunction...) inspm_script.mto replace the linepreproc_data = spm_preprocessing(f_fn, s_fn, fwhm, spm_dir);. You would, of course, need to make sure that variables names are correct and don’t interact badly with variables already defined in the script.
I would recommend option 2, even if it’s a bit of work to merge the function within your script, because you don’t have to compile SPM yourself.
As I mentioned before, you can also pass input arguments from the command line to your script using the inputs variable. For example, if spm_script.m contains:
directory = inputs{1};
subject = inputs{2};
fprintf('Preprocessing %s %s\n',directory,subject);
you can execute:
./run_spm12.sh /usr/local/MATLAB/MATLAB_Runtime/v95/ script spm_script.m /data subj-01
Thank You so much Guillaume. You helped me a lot. And now code is work by using method 2. I have copied all code of spm_preprocessing.m except first line that is
function output = spm_preprocessing(functional4D_fn, structural_fn, fwhm, spm_dir)
It running but at the end one error occurs after step 5 done
Step 5 - done!
Undefined variable "preproc_data" or class "preproc_data.rf_fn".
Error in spm_script (line 228)
spm_check_registration(s_fn, [preproc_data.rf_fn ',1'], preproc_data.rgm_fn, preproc_data.rwm_fn, preproc_data.rcsf_fn)The code in spm_preprocessing defines a variable output while the rest of the code in spm_script expects preproc_data. Either replace all instances of output by preproc_data or add this line at the right place:
preproc_data = output;