Hi Johannes, yes you are right, the outputspec is missing, that can be added similar to the inputspec…
yes, to compile you need MATLAB Compiler (mcc), to get your matlab scritps into a standalone application that would only require matlab runtime (MCR) of the same version thru which you compiled your matlab code.
Hi, I am working on spm12 and I want to run my scripts using spm12 functions on a Linux server using commands. I am facing some errors. Do you have any idea to run these scripts on a Linux server using the MCR installer.
Hi,
Have you got the standalone version of spm12? if yes, then you can run your .m scripts with spm12 and MCR as one of the following commands:
./run_spm12.sh /path/to/MCR/vX script your_script.m
your_script.m should be calling the spm functions.
cheers…
will it work for the Linux server? without GUI
Where to got spm12 standalone for R2018b mcr. If not available can you please guide me how to create SPM12 Standalone Application?
A version of SPM12 standalone for MATLAB Runtime R2019b is available here (temporary link). The corresponding MATLAB Runtime itself can be downloaded here.
If you want to compile SPM yourself, instructions are available here (third item in the list).
Yes, it should, as mentioned by @mohammad.shahid. Could you copy-paste here the error messages you observed?
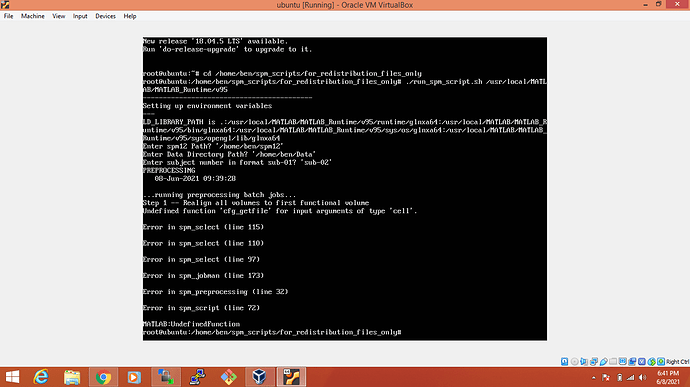
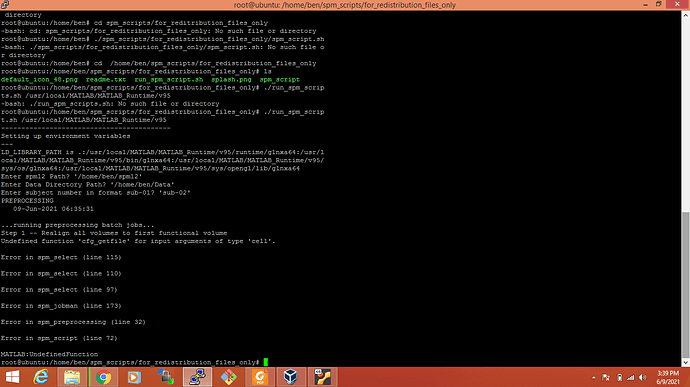
Ok. First, let me explain to you what I want actually. I have created a script that takes data and spm12 path and calls a function that is also created by me names as preprocessing. In this function, I have called spm12 functions for preprocessing of data. I have created a standalone application as ssh to run these scripts on the Linux server and used the spm12 standalone version. It’s showing errors like this:
Thank you - this is useful.
The error about cfg_getfile indicates that you compiled SPM yourself without relying on spm_make_standalone: using this function should solve the issue for you.
Note that when using the syntax script your_script.m, you can pass parameters from the command line and they will be available from a variable called inputs (a cell array) in your script. This means you could pass the data directory and the subject index from the command line with any interactive step. Also you shouldn’t have to provide the location of the SPM installation - if you need this information from a script, using spm('Dir').
Finally, concerning the error about spm_preprocessing: this is not an SPM function so, if it is a function (and not a script), make sure you include it in your SPM compilation.
I have used spm_make_standalone function for SPM compilation and there was an option to add required files to run…
What to add there or need to left that empty??
I want to remove the error about cfg_getfile
I am frustrated. I want to run my standalone application of .m scripts using spm12 on linux server. Is there any easier way to tackle first error about ‘cfg_getfile’
Which option from spm_make_standalone are you referring to? The compiler is configured to include all SPM files.
If you only one to execute scripts (i.e. not defining new functions) then you don’t need to compile yourself and can use the standalone SPM linked above.
I need SPM12 with MCR 2018b. Let me try to compile by your method then I’ll let you know. Thanks Alot
OK, let me know how it goes. If you give me a bit of time, I could compile one version for you with R2018b. Searching through old files, I could find this (r7487, R2018b).
I also have used this r7487, and thay cfg_getfile error was using this one. And yes if you can compile one version for me I’ll be very very thankful to you. I am using R2018b MCR and Matlab for Linux
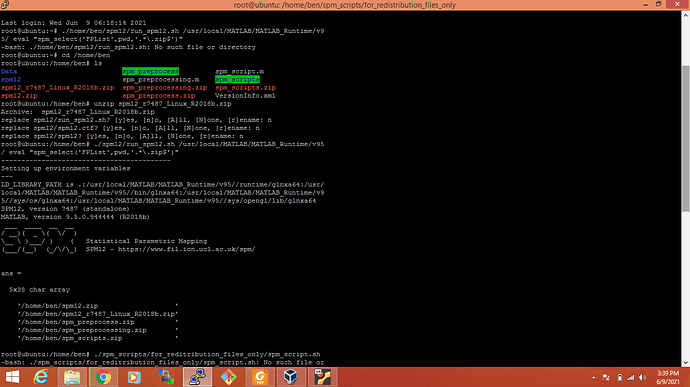
I am surprised you got the cfg_getfile error with that version. I tried installing and using it myself and it seems to work fine:
wget https://www.fil.ion.ucl.ac.uk/spm/download/restricted/utopia/dev/soon_gone/spm12_r7487_Linux_R2018b.zip
wget https://ssd.mathworks.com/supportfiles/downloads/R2018b/deployment_files/R2018b/installers/glnxa64/MCR_R2018b_glnxa64_installer.zip
unzip MCR_R2018b_glnxa64_installer.zip
sudo ./install -mode silent -agreeToLicense yes
unzip spm12_r7487_Linux_R2018b.zip
./spm12/run_spm12.sh /usr/local/MATLAB/MATLAB_Runtime/v95/ eval "spm_select('FPList',pwd,'.*\.zip$')"
which will display:
SPM12, version 7487 (standalone)
MATLAB, version 9.5.0.944444 (R2018b)
___ ____ __ __
/ __)( _ \( \/ )
\__ \ )___/ ) ( Statistical Parametric Mapping
(___/(__) (_/\/\_) SPM12 - https://www.fil.ion.ucl.ac.uk/spm/
ans =
2x58 char array
'/tmp/R2018b/MCR_R2018b_glnxa64_installer.zip'
'/tmp/R2018b/spm12_r7487_Linux_R2018b.zip '
Doest this work for you?
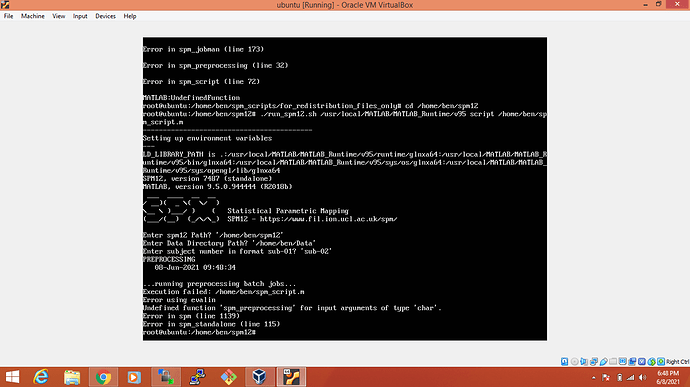
Worked. SPM is working. But while executing the shell script of my code that I want to run on Linux server. The same
code is working properly on windows and Linux DesktopProblem is same
I think you need to be a bit more explicit of what you are doing with your scripts. From what I see on the screenshots, it looks like you are trying to compile them? It seems that you have two *.m files, spm_script.m and spm_preprocessing.m: are they scripts or functions (starting with function spm_*)? Best would be to have a single script, that therefore does not need to be compiled and that you can directly provide to the SPM standalone command line:
./spm12/run_spm12.sh /usr/local/MATLAB/MATLAB_Runtime/v95/ script spm_script.m