I have a statistical image where the value of each parcel corresponds to a t-value from a one-sample t-test. In other words: Each value represents the deviation from zero (so it’s not a contrast image, that for example compares two conditions or two groups of subjects like in a second-level-analysis). Now I would like to extract regions of interest from that image with particularly high values compared to the rest of the brain. This is how my image looks like:
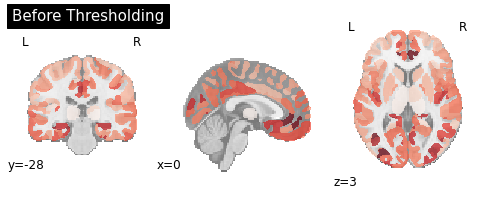
You can visually see that there are peaks around the anterior and posterior cingulate cortex and in the insula. and I would intuitively expect most of these regions to survive the thresholding whilst all other regions are set to zero. I also have p-values for my t-values but all of these are extremely low so even after using a multiple comparison correction with a very low alpha everything stays significant. I somehow need a method that is agnostic to p-values and simply works with the parcel-values. Already thought about Otsu-Tresholding, but I am not sure if I am on the right path here.
@bthirion : Do you have an idea how to solve this?