Hi @ChrisGorgolewski,
Okay, thanks just wanted to make sure.
I followed your suggestion regarding how to move the mask from MNI to T1w space:
Here’s how I did it in a Nipype script:
ApplyTransforms Node
from nipype.interfaces.ants import ApplyTransforms
transform = pe.Node(interface=ApplyTransforms(), name='transform')
transform.inputs.dimension = 3
transform.inputs.float = False
transform.inputs.interpolation = 'NearestNeighbor'
Part of workflow
wf.connect(selectfiles, 'vismask', transform, 'input_image')
wf.connect(selectfiles, 'transforms', transform, 'transforms')
wf.connect(selectfiles, 'anat', transform, 'reference_image')
wf.connect(transform, 'output_image', datasink, 'transform')
where vismask is the mask over visual cortex in MNI space, transforms is the *.h5-file and the reference_image is the subject’s T1w image. This is what the executed command looks like:
antsApplyTransforms --default-value 0 --dimensionality 3 --float 0 \
--input /path/to/mask/vismask.nii --interpolation Linear --output vismask_trans.nii \
--reference-image /project/derivatives/fmriprep/sub-01/anat/sub-01_desc-preproc_T1w.nii.gz \
--transform /project/derivatives/fmriprep/sub-01/anat/sub-01_from-MNI152NLin2009cAsym_to-T1w_mode-image_xfm.h5
So it works. I get a mask that is reasonably transformed into subject space (Note: The mask in MNI space still needs to be improved and obviously it not only covers visual cortex…):
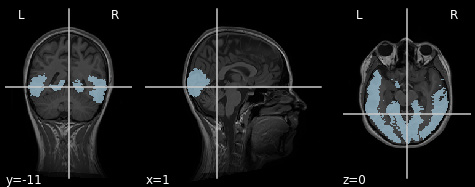
Anyway, now I have yet another problem: When I load the functional data into the mask in Nilearn it takes very long (which is okay, since it’s a lot of data) but eventually crashes. I use the NiftiMasker like this:
masker = NiftiMasker(mask_img = path_mask)
masked_data = masker.fit_transform(path_func)
I don’t get this problem when I use the FMRIPREP output *space-T1w_desc-brain_mask.nii.gz as a “T1w whole brain mask”.
Any ideas? Thanks!

