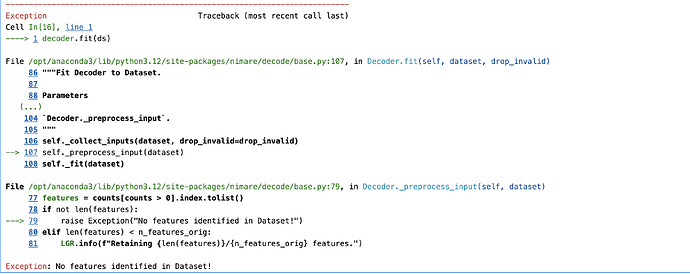
Hello everyone,
I am currently using NiMARE to decode my ALE results. For example, after completing the meta-analysis, I obtained activation results in the Frontal pole (x, y, z: -6, 58, 18). I created a spherical ROI with the following code:
from nilearn import datasets, plotting
from nilearn.input_data import NiftiSpheresMasker
from nilearn.maskers import _unmask_3d
import nibabel as nib
from nibabel import Nifti1Image
# Create spherical ROI
brain_mask = datasets.load_mni152_brain_mask()
_, A = NiftiSpheresMasker._apply_mask_and_get_affinity(
seeds=[(-6, 58, 18)],
niimg=None,
radius=10,
allow_overlap=False,
mask_img=brain_mask)
FPole_mask = _unmask_3d(
X=A.toarray().flatten(),
mask=brain_mask.get_fdata().astype(bool))
FPole_mask = Nifti1Image(FPole_mask, brain_mask.affine)
nib.save(FPole_mask, "FPole.nii.gz")
# Plot the result to make sure it makes sense
plotting.plot_roi("FPole.nii.gz")
plotting.show()
roi_img = nib.load("FPole.nii.gz")
Then, I used NiMARE to decode my image:
import nimare
# Fetch Neurosynth data (Note: This can take a while!)
databases = nimare.extract.fetch_neurosynth(data_dir='../data')[0]
# Convert to NiMARE dataset (Note: This can take a while!)
ds = nimare.io.convert_neurosynth_to_dataset(
coordinates_file=databases['coordinates'],
metadata_file=databases['metadata'],
annotations_files=databases['features']
)
# Perform decoding
decoder = nimare.decode.discrete.ROIAssociationDecoder(roi_img)
decoder.fit(ds)
decoded_df = decoder.transform()
print(decoded_df.iloc[60:80, :].to_string())
Plotted the decoding result with the following code:
import pandas as pd
import matplotlib.pyplot as plt
from wordcloud import WordCloud
# Save decoding result
decoded_df.to_csv('FPole_decoded_output.csv', index=True)
# Generate word cloud
word_path = "FPole_decoded_output.csv"
data1 = pd.read_csv(word_path)
word_freq = dict(zip(data1['feature'], data1['r']))
wordcloud = WordCloud(width=800, height=400, max_font_size=100, background_color='white').generate_from_frequencies(word_freq)
plt.figure(figsize=(10, 5))
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
I have the following questions:
- Is my decoding approach correct? If it is correct, are there other visualization methods I can use?
- The results include all terms, but in my research, I am only interested in cognitive-related terms. I discovered that the Cognitive Atlas only includes cognitive vocabulary, but I don’t know how to replace the vocabulary in NiMARE. I also noticed that if I decrease the number of terms, the model’s fit worsens. For example, I don’t care about anatomical terms. How can I exclude these irrelevant terms from the results? How can this be achieved in Python?
Thank you very much for your help!