I am trying to run ICA-FIX on fMRIPrep outputs. Example of a converted directory:
Permissions Size User Date Modified Name
drwxrwxr-x - mpasternak 6 Mar 16:01 .
drwxrwxr-x - mpasternak 2 Mar 21:01 ├── filtered_func_data.ica
.rw-rw-r-- 2.7k mpasternak 2 Mar 10:28 │ ├── eigenvalues_percent
.rw-rw-r-- 891 mpasternak 2 Mar 21:01 │ ├── hand_labels_noise.txt
.rw-rw-r-- 5.1k mpasternak 2 Mar 10:29 │ ├── log.txt
.rw-rw-r-- 21k mpasternak 2 Mar 10:28 │ ├── mask.nii.gz
.rw-rw-r-- 991k mpasternak 2 Mar 10:28 │ ├── mean.nii.gz
.rw-rw-r-- 112k mpasternak 2 Mar 10:28 │ ├── melodic_dewhite
.rw-rw-r-- 50k mpasternak 2 Mar 10:28 │ ├── melodic_FTdewhite
.rw-rw-r-- 48k mpasternak 2 Mar 10:28 │ ├── melodic_FTmix
.rw-rw-r-- 39M mpasternak 2 Mar 10:28 │ ├── melodic_IC.nii.gz
.rw-rw-r-- 2.0k mpasternak 2 Mar 10:28 │ ├── melodic_ICstats
.rw-rw-r-- 105k mpasternak 2 Mar 10:28 │ ├── melodic_mix
.rw-rw-r-- 39M mpasternak 2 Mar 10:28 │ ├── melodic_oIC.nii.gz
.rw-rw-r-- 39M mpasternak 2 Mar 10:29 │ ├── melodic_pca.nii.gz
.rw-rw-r-- 19 mpasternak 2 Mar 10:28 │ ├── melodic_pcaD
.rw-rw-r-- 615k mpasternak 2 Mar 10:28 │ ├── melodic_pcaE
.rw-rw-r-- 17k mpasternak 2 Mar 10:28 │ ├── melodic_PPCA
.rw-rw-r-- 105k mpasternak 2 Mar 10:28 │ ├── melodic_Tmodes
.rw-rw-r-- 121k mpasternak 2 Mar 10:28 │ ├── melodic_unmix
.rw-rw-r-- 115k mpasternak 2 Mar 10:28 │ ├── melodic_white
.rw-rw-r-- 979k mpasternak 2 Mar 10:28 │ ├── Noise__inv.nii.gz
drwxrwxr-x - mpasternak 2 Mar 10:29 │ └── report
.rw-rw-r-- 202M mpasternak 2 Mar 10:29 ├── filtered_func_data.nii.gz
drwxrwxr-x - mpasternak 6 Mar 15:56 ├── fix
.rw-rw-r-- 21k mpasternak 2 Mar 10:28 ├── mask.nii.gz
.rw-rw-r-- 28k mpasternak 2 Mar 10:25 ├── mask_non_dilated.nii.gz
drwxrwxr-x - mpasternak 2 Mar 10:24 ├── mc
.rw-rw-r-- 12k mpasternak 2 Mar 10:24 │ ├── prefiltered_func_data_mcf.par
.rw-rw-r-- 269k mpasternak 2 Mar 10:24 │ └── prefiltered_func_data_mcf.png
.rw-rw-r-- 993k mpasternak 2 Mar 10:28 ├── mean_func.nii.gz
.rw-rw-r-- 640 mpasternak 6 Mar 15:56 ├── pyfix.log
drwxrwxr-x - mpasternak 6 Mar 16:01 └── reg
.rwxrwxrwx 2.9M mpasternak 27 Jan 18:09 ├── example_func.nii.gz
.rwxrwxrwx 8.4M mpasternak 6 Mar 15:50 ├── highres.nii.gz
.rw-rw-r-- 144 mpasternak 2 Mar 10:24 └── highres2example_func.mat
I encounter the following error when running fix -f . as seen in the log file:
15:56:53 INFO fix.py 447: pyfix 0.9.0
15:56:53 INFO fix.py 448: date: 2025-03-06 (Thursday)
15:56:53 INFO fix.py 449: pwd: /home/mpasternak/Documents/MPasternak_fMRI_BOLD_Test/derivatives/fsl/sub-GRN225/ses-V11
15:56:53 INFO fix.py 450: command: .
15:56:53 INFO extract.py 265: Creating structural segmentation with FSL FAST
15:56:53 DEBUG run.py 363: Calling command: /home/mpasternak/fsl/bin/fast --out=fix/fastsg -t 1 reg/highres.nii.gz
15:56:55 DEBUG run.py 389: Standard output: Exception: Not enough classes detected to init KMeans
I don’t understand why FAST is failing with this KMeans business. The highres image should be perfectly compatible, as it requires the image to be brain-extracted:
Any idea what troubleshooting I can do to?
Hi @Maurice_Pasternak, that error usually indicates that there is something about the voxel intensities in the T1 image which is preventing FAST from properly modelling the different tissue classes. This can happen for a range of reasons on preprocessed T1s, for example it is quite common for fast to fail if the tissue intensities are too homogenous. If you can share your T1 image with me, I might be able to suggest something. Alternatively, you could try running fast manually before calling fix - it won’t re-run fast if the fastsg_* files are already present in the fix/ directory.
Thanks @paulmccarthy for the suggestion!
Please find the scans here: FAST_Troubleshooting – Google Drive
- The
*desc-preproc_T1w.nii.gz image is a pre-processed native space T1w image
- The
*.h5 file is the forward transformation
The other two files are based off using antsApplyTransforms on the aforementioned T1w image. They only differ in one aspect; the reference used.
- For the
res-1 image, I used the tpl-MNI152NLin6Asym_res-01_desc-brain_T1w.nii.gz provided by Templateflow
- For the
res-T1w image, I resampled the tpl-MNI152NLin6Asym_res-01_desc-brain_T1w.nii.gz image to have the same zooms/resolution as the native-space T1w image (1.0156, 1.0156, 1.0156) and used this resampled image as the reference. I used this Nibabel function to accomplish the resampling.
My observations:
- The
res-1 image fails FAST while the res-T1w image does not.
- For other subjects in my dataset the
res-1 image may sometimes succeed. The res-T1w image consistently succeeds…for now.
This is still curious to see, as I can’t image a difference in resolution of 0.0156mm is enough to cause such a drastic difference.
Hi @Maurice_Pasternak, it’s not clear to me which image you have used as the reg/highres.nii.gz that you pass to fast/fix. This image should be the native space T1 to which the filtered_func_data.nii.gz file is registered (via reg/example_func2highres.mat), but it sounds like you’re using a copy which has been resampled into the standard template space? Why aren’t you using (a brain-extracted version of) the *desc-brain_T1w image?
Long story short, I am working off the outputs of fMRIPrep in which the preprocessed BOLD data (realigned, unwarped, slice-time-corrected, etc.) was only output in standard space. Therefore, I’m working around that constraint. While fMRIPrep did output an intermediate native space preprocessed T1w image, I can’t use it with FAST since it may not align with the BOLD data.
Yes, I do realize this goes against FSL convention of operating on partially preprocessed native space data with ICA-FIX, then transforming the denoised data to standard space.
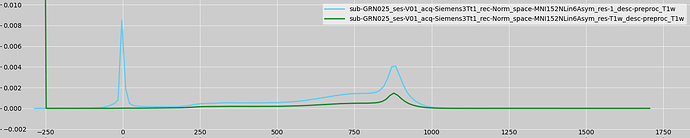
I am using the *_space-MNI152NLin6Asym_res-T1w_desc-preproc_T1w.nii.gz (standard space, maintain original 1.0156mm resolution) image as the reg/highres.nii.gz input, as I found that it is the only standard space structural image that will be accepted by FAST. What I don’t understand is why this is the case that is compatible, as the distribution of pixel intensities should not differ drastically between 1mm and 1.0156mm resolution versions of the same T1w scan.
On the contrary, the distribution of voxel intensities in those two images appears to be quite different:
I’m not a FAST expert so I can’t give you an explanation as to why it behaves differently on these images. But I know from experience that it is sensitive to these kinds of things. It was written to expect a T1 image with a “typical” distribution of voxel intensities. For example, I doubt that negative values were ever considered, because they will never occur in a normal T1 which has not been resampled/interpolated.
Why not just use the native space T1, and adjust the func<->struct<->standard transformations accordingly?