I am conducting task-based fMRI analyses using fmriprep and nipype.
After preprocessing the data, I am cleaning the data in NiPype as follows:
confound_df = pd.read_csv(confound_file, delimiter="\t")
# Motion outliers (fd threshold from fmriprep defaults)
motion_outliers = [col for col in confound_df.columns if "motion_outlier" in col]
# PCA first 5 components
# a_comp_cor = [
# "a_comp_cor_00",
# "a_comp_cor_01",
# "a_comp_cor_02",
# "a_comp_cor_03",
# "a_comp_cor_04",
# "a_comp_cor_05",
#]
a_comp_cor = []
# In case of dummy scans
non_steady_state = [
col for col in confound_df.columns if col.startswith("non_steady_state")
]
# Motion regressors
motion = ["trans_x", "trans_y", "trans_z", "rot_x", "rot_y", "rot_z"]
fd = ["framewise_displacement"]
# Combine them all
filter_col = np.concatenate(
[motion_outliers, a_comp_cor, non_steady_state, motion, fd]
)
confound_df = confound_df[filter_col]
# Fill nans with 0's
confound_df = confound_df.fillna(0)
if verbose:
print(f"Confounds: {confound_df.columns}")
confounds_matrix = confound_df.values
confounds_output_file = get_output_name(confound_file, output_path, "_trimmed.csv")
np.savetxt(confounds_output_file,confounds_matrix,delimiter=",")
# Load in functional data
raw_func_img = nimg.load_img(func_file)
# Make sure these are the same length.
assert raw_func_img.shape[3] == confounds_matrix.shape[0]
# Clean!
clean_img = nimg.clean_img(
raw_func_img,
confounds=confounds_matrix,
detrend=True,
standardize=True,
low_pass=LOW_PASS,
high_pass=HIGH_PASS,
t_r=T_R,
mask_img=mask_file,
)
Strangely, (for at least two subjects) this is resulting in a NEGATIVE correlation between the raw timeseries and the cleaned timeseries (for a few random ROIs).
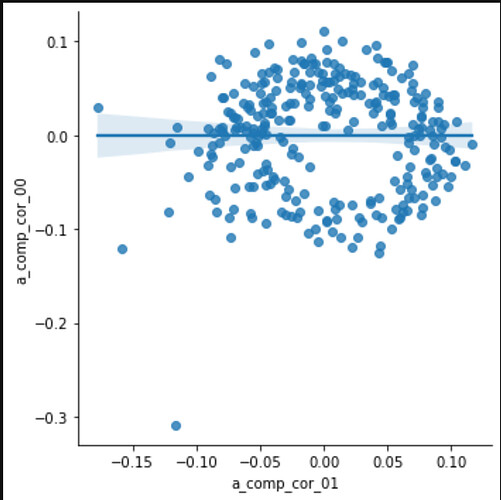
Also strangely, the first two components plotted against each other make a circle?
Does anyone have ideas what might be causing this? Are some of my other confounds oddly redundant? (All of the a_comp_cors are ‘combined’ according to the json).