Summary of what happened:
I’m trying to extract the mean timeseries from a masked region but having issues with the mask. The functional data was preprocessed with the default fMRIPrep settings and is in MNI152 space with 3mm isometric dimensions. The masks were pulled from the SUIT atlas, are in MNI152 space (though the header via fslhd has qform_name as “Unknown”), but have 1mm iso resolution.
Question:
How do I downsample this mask to a compatible 3mm resolution without moving the mask?
Solutions Edit:
This following solution worked for me and was found in the FSL archives (JISCMail - FSL Archives):
flirt -in ORIGINAL_MASK.nii.gz \
-ref FEAT_DIRECTORY/example_func.nii.gz \
-out FEAT_DIRECTORY/NEW_MASK.nii.gz \
-applyxfm -usesqform
However, check out @Steven 's solution below which takes a much simpler path just using Nilearn.
What I’ve Tried:
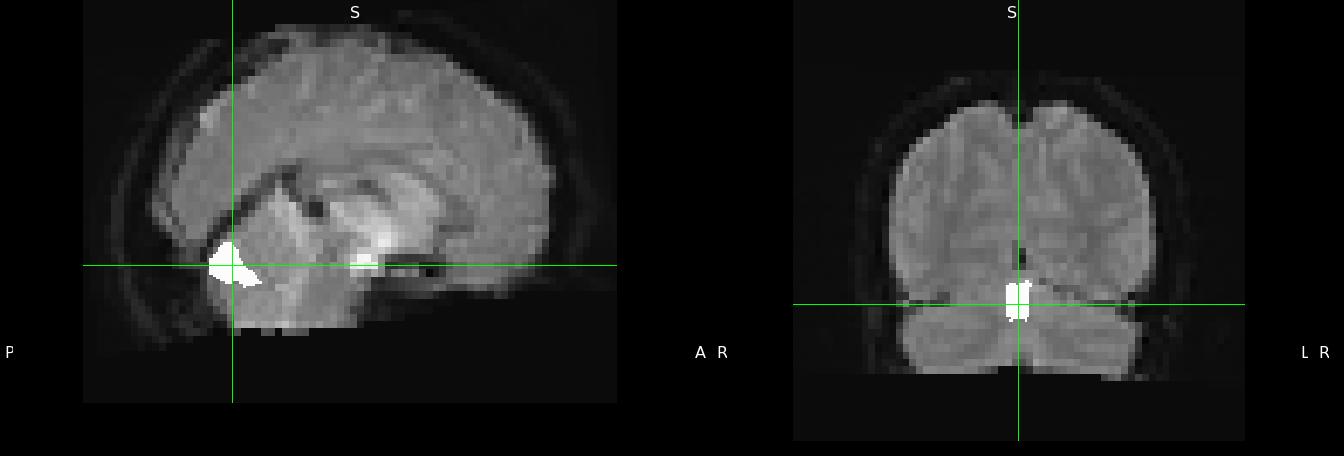
This is an example of a Vermis mask and its placement is right where we need it:
But the dimension differences generate this error with fslmeants:
ERROR: Mask and Input volumes have different (x,y,z) size.
fslmeants -i ${DERIV}/${subj}/func/${subj}_task-contact_run-${run}_space MNI152NLin2009cAsym_desc-preproc_bold_smooth6mm.nii.gz \
-o ${DERIV}/${subj}/func/${subj}_task-contact_run-${run}_roi-VermisVI_timecourse.txt \
-m ${ROIPATH}/VermisVI_bin_3mm.nii.gz
Where $DERIV is the filepath to our subject data, $ROIPATH is a directory to our mask nifti files, and $subj and $run are variables I iterate through in a for loop. I’ve tried downsampling the mask to 3mm resolution using flirt like this:
flirt -in ${ROIPATH}/VermisVI_bin.nii.gz \
-ref ${FEAT}/example_func.nii.gz \
-out ${FEAT}/rois/${subj}_VermisVI_func \
-applyxfm -init ${FEAT}/reg/example_func2standard.mat
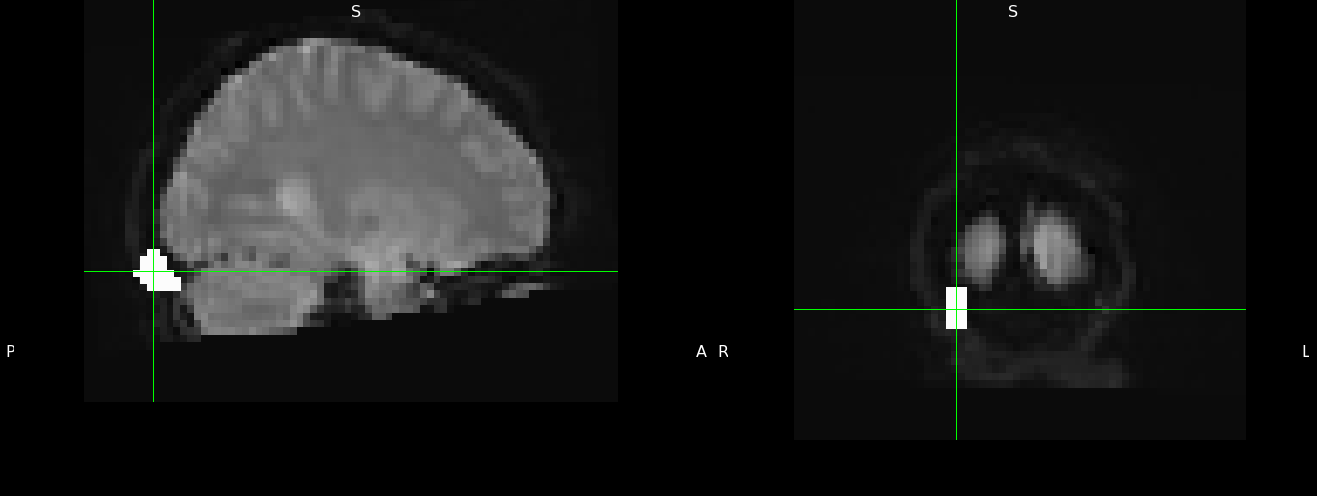
Where $FEAT is a filepath to a first level FEAT directory which autogenerated the example_func file. The example_func2standard.mat was pulled from the FSL directories according to the Jeanette Mumford workaround. This makes the mask compatible, but it inevitably moves the mask off the brain:
I’ve tried resampling as suggested in Andy’s Brain Blog and the FSL Archives:
flirt -in ${ROIPATH}/VermisVI_bin.nii.gz \
-ref ${ROIPATH}/VermisVI_bin.nii.gz \
-applyisoxfm 3.0 -nosearch \
-out ${ROIPATH}/VermisVI_bin_3mm.nii.gz
fslhd showed that the various pixdim's update to 3.0mm but the other details (i.e., dim, qto_xyz, sto_xyz) don’t seem to change, resulting in the previously noted error.
Version:
Version 6.0 of FSL; Version of fMRIPrep unknown but could find out if needed.
Additional Information:
An added complication is that I don’t have direct access to this data. I’m advising a student in another lab who is working on it. As such avoiding re-processing the data, if at all possible, would be ideal. I’ve made the original mask available here if it’s helpful: VermisVI.nii.gz - Google Drive